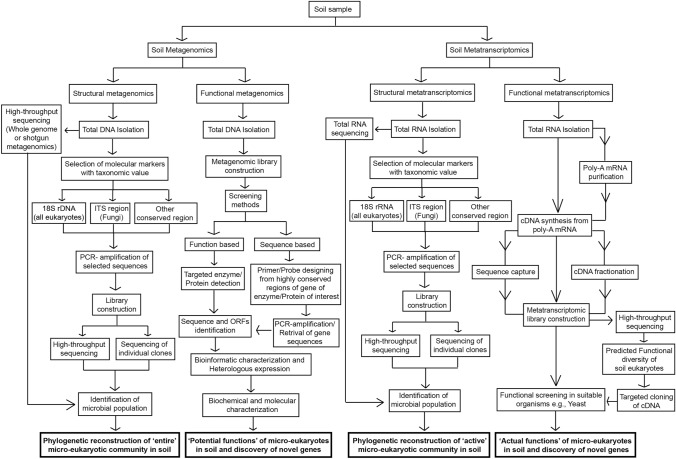
Fig. 1.
Cumulative description of processes and pipelines of metagenomics and metatranscriptomics to study soil micro-eukaryotes. Flow diagram is showing DNA-based metagenomic and RNA-based metatranscriptomic approaches to study community profiles (structural) and functions (functional) of micro-eukaryotes in the soil. Structural metagenomics employs sequencing of metagenomic or PCR-amplified marker DNA for the study of entire micro-eukaryotic community structure in the soil. However, structural metatranscriptomics is based on sequencing of total RNA- or PCR-amplified cDNA synthesized from marker RNA sequence for the study of active micro-eukaryotic community in the soil. Functional metagenomics includes metagenomic library construction followed by function- or sequence-based study of individual clones. This library is used to decipher potential functions of micro-eukaryotes in the soil and screening of genes encoding enzymes/proteins of interest. Functional metatranscriptomics relies on cDNA library constructed from soil-extracted micro-eukaryotic mRNA. This library is further used for the study of actual functional diversity of soil micro-eukaryotes through high-throughput sequencing and in silico functional predictions of sequences. This metatranscriptomic library is also exploited for the screening of micro-eukaryotic genes with desired function by expressing them in a suitable expression system. Studies based on the described approaches are being applied to explore the community structure of soil micro-eukaryotes and to screen novel functional eukaryotic genes that may have applications in biotechnology