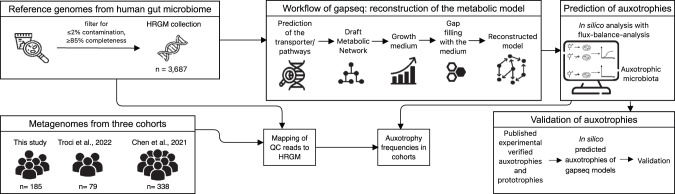
Fig. 1. Workflow for the prediction of auxotrophies with genome-scale metabolic modeling.
Gapseq was used to reconstruct genome-scale metabolic models from genomes of the Human Reference Gut Microbiome (HRGM) catalog [27]. The workflow of gapseq to reconstruct metabolic models consists of five steps: transporter/metabolic pathway prediction, draft metabolic network construction, growth medium prediction, gap filling, final model reconstruction. Auxotrophy prediction was performed using flux-balance analysis and validated by reconstructing gapseq models from experimentally verified auxotrophic strains. The predicted auxotrophies were compared on strain level from gapseq and AGORA2 models to experimentally verified auxotrophies. QC reads of cohorts were mapped on HRGM. Auxotrophy frequencies in cohorts were determined by mapping QC reads from the metagenomes of the cohorts to genomes from HRGM collection. Icons are from www.flaticon.com (creators: photo3idea_studio, Freepik, surang, Eucalyp, Voysla, juicy_fish, smashingstocks, SBTS2018, creative_designer).