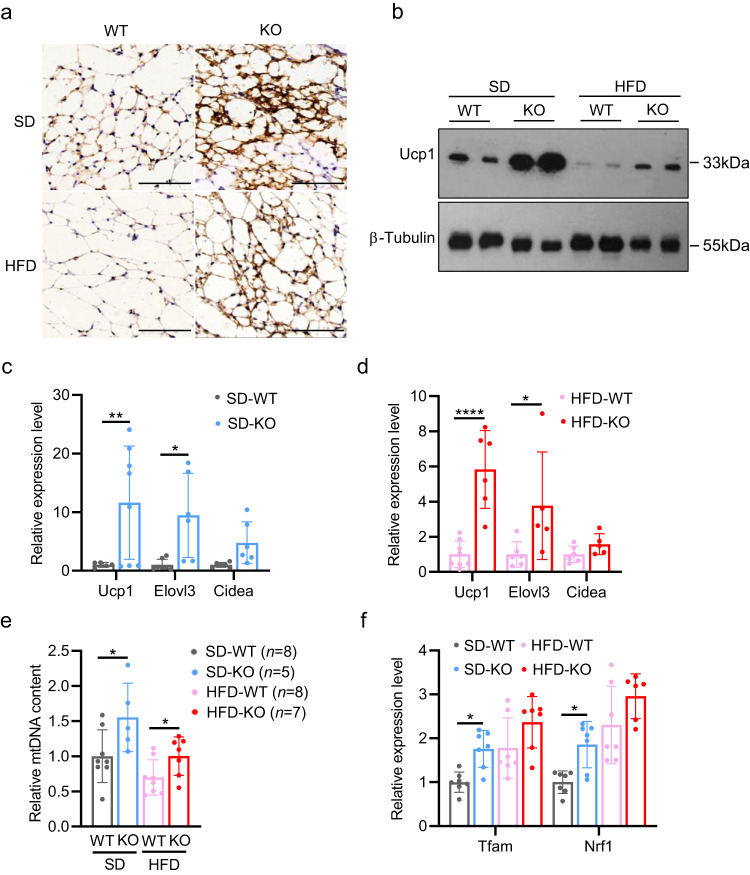
Fig. 3. iWAT browning occurs in Myod KO mice.
a Representative immunostaining of uncoupling protein 1 (Ucp1) on cryosections of iWAT from Myod KO mice and WT littermates fed with SD or HFD, n = 3 mice. Scale bar, 100 μm. b Western blots showing protein levels of Ucp1 in iWAT from Myod KO mice and WT littermates fed with SD or HFD. c Relative mRNA levels of browning-related genes in iWAT from Myod KO mice and WT littermates fed with SD, as determined by RT-qPCR. For Ucp1 gene, n = 5 (SD-WT), n = 8 (SD-KO) mice. p = 0.0061 (SD-WT vs. SD-KO); For Elovl3 gene, n = 7 (SD-WT), n = 6 (SD-KO) mice. p = 0.0293 (SD-WT vs. SD-KO); For Cidea gene, n = 6 (SD-WT), n = 6 (SD-KO) mice. d Relative mRNA levels of browning-related genes in iWAT from Myod KO mice and WT littermates fed with HFD, as determined by RT-qPCR. For Ucp1 gene, n = 8 (HFD-WT), n = 6 (HFD-KO) mice. p < 0.0001 (HFD-WT vs. HFD-KO); For Elovl3 gene, n = 6 (HFD-WT), n = 5 (HFD-KO) mice. p = 0.0173 (HFD-WT vs. HFD-KO); For Cidea gene, n = 6 (HFD-WT), n = 5 (HFD-KO) mice. e Quantification of mitochondrial DNA content in iWAT from Myod KO mice and WT littermates fed with HFD or SD, p = 0.0412 (SD-WT vs. SD-KO), p = 0.0430 (HFD-WT vs. HFD-KO). f Relative mRNA levels of mitochondrial biogenesis-related genes in iWAT from Myod KO mice and WT littermates fed with SD or HFD, as determined by RT-qPCR. For Tfam gene, n = 7 (SD-WT), n = 7 (SD-KO), n = 7 (HFD-WT), n = 7 (HFD-KO) mice. p = 0.0354 (SD-WT vs. SD-KO). For Nrf1 gene, n = 7 (SD-WT), n = 7 (SD-KO), n = 7 (HFD-WT), n = 6 (HFD-KO) mice. p = 0.0151 (SD-WT vs. SD-KO). Data are presented as mean ± SD. Significance was assessed by two-way ANOVA (c-d, f) or two tail Student’s t-test (e). *p < 0.05, **p < 0.01, ****p < 0.0001 compared to WT control group. Source data are provided as a Source Data file.