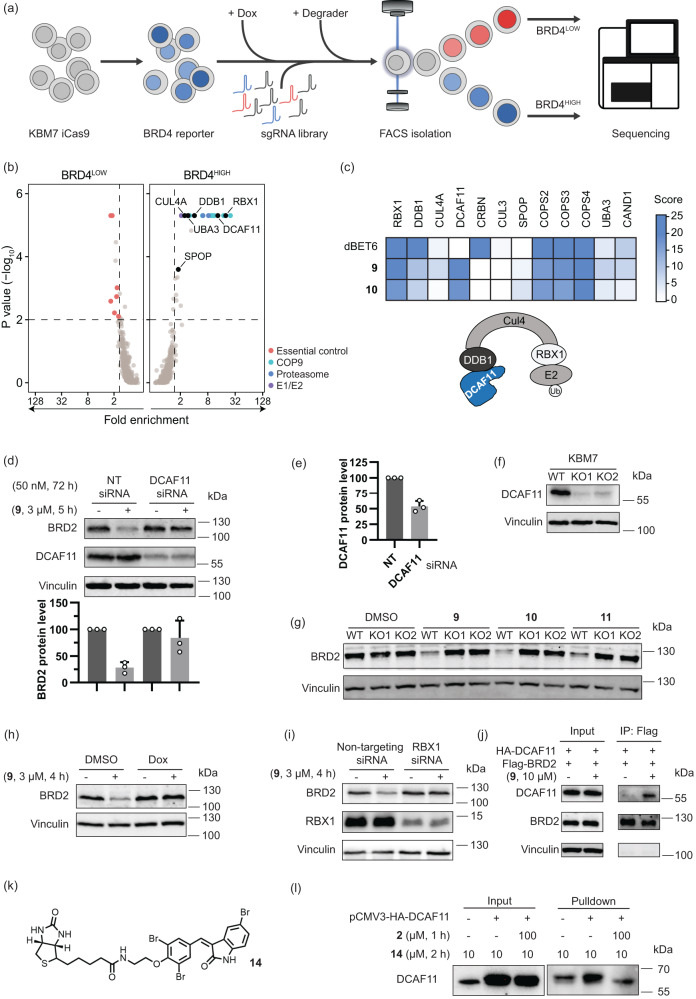
Fig. 3. Compound 9 promotes proteasomal degradation of BET proteins via the CRLDCAF11 complex.
a Schematic of FACS-based BRD4 protein stability CRISPR screen assay. b, c Identification of genes required for compound 9-mediated degradation of BRD4. b FACS-based CRISPR screens for regulators of BRD4 degradation induced by 9. Average gene-level fold-changes and non-adjusted one-sided p-values of BRD4HIGH and BRD4LOW cell populations compared to BRD4MID fraction were calculated using MAGeCK46. Essential control genes (BRD4LOW) and 20S proteasome subunits, COP9 signalosome subunits and E1 or E2 ubiquitin ligases (BRD4HIGH) inside the scoring window (p-value < 0.01, fold-change > 1.5) are labelled. c Heatmap of selected screen hits from Fig. 3b and Supplementary Fig. 4a, b. Gene-level score was calculated as Log2 (fold change) * Log10 (p-value). d, e BRD2 and DCAF11 protein levels in Hep G2 cells transfected with non-targeting (NT) siRNA or DCAF11 siRNA, followed by 9 addition. d Quantification of the relative BRD2 levels. Data are mean values ± SD (n = 3 biological replicates). e Quantification of the relative DCAF11 levels. Data are mean values ± SD (n = 3 biological replicates). f DCAF11 levels in wild type (WT) KBM7 cells and DCAF11 knockout (KO1 and KO2) KBM7 cells. Representative result of n = 3. g BRD2 levels in WT and DCAF11 knockout (KO1 and KO2) KBM7 cells treated with compound 9 at 3 μM, 10 at 10 μM and 11 at 10 μM, for 6 h. Data are representative of n = 3. h BRD2 levels in Dox-inducible DDB1 knockout KBM7 cells pretreated with 0.5 µg/mL Dox for 3 days followed by addition of 9 at 3 μM for another 4 h. Representative result of n = 3. i BRD2 and RBX1 levels in MDA-MB-231 cells with NT or RBX1 siRNA followed by 9 addition. Representative result of n = 3. j BRD2 co-immunoprecipitation. FLAG-BRD2 protein was enriched by anti-FLAG beads and samples were analyzed for DCAF11. Representative result of n = 2. k Structure of biotinylated affinity probe 14. l Affinity-based enrichment by pulldown in Hep G2 cells overexpressing DCAF11. Protein levels were analyzed by WB. Representative result of n = 2.