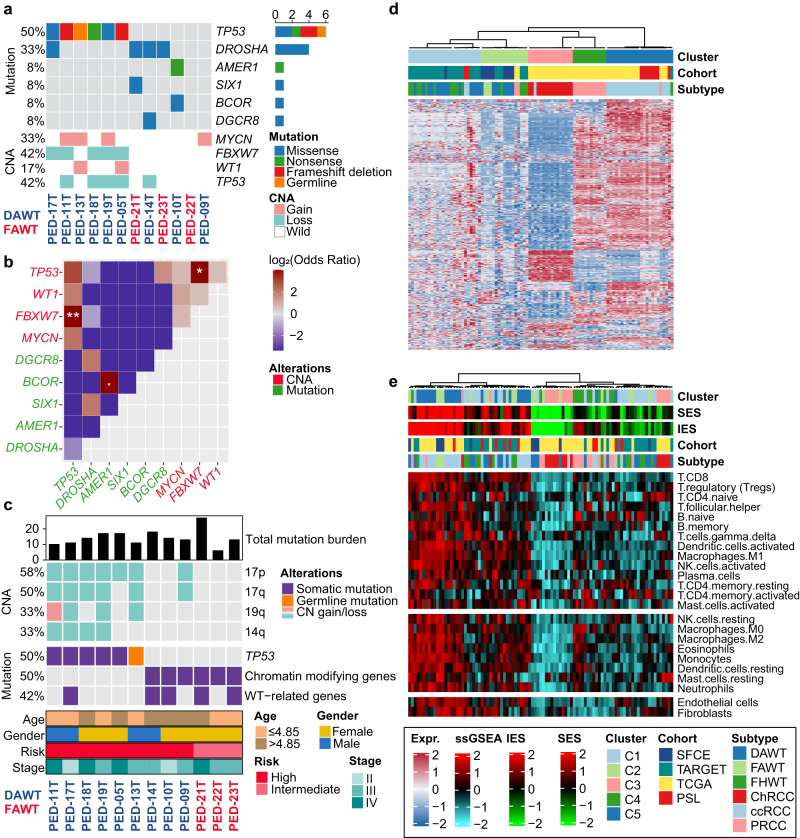
Fig. 1. Genome-wide landscape of 12 Wilms tumors (WTs) identified by whole-exome sequencing and transcriptome pattern among 95 representative patients with four types of kidney cancers.
a OncoPrint showing the mutation and copy number alteration (CNA) of Wilms tumor-related genes. b Genetic mutual exclusivity or cooccurrence in the SFCE-WT cohort using a one-side Fisher’s exact test (. P < 0.1, * P < 0.05, ** P < 0.01). c OncoPrint showing broad CNA (>25%) and mutations in a selection of frequently mutated genes arranged vertically by functional group. Total mutation burden was annotated at the top panel, and clinicopathological information was annotated in the bottom panel. d Heatmap showing the distinct expression pattern of four kidney cancers by unsupervised consensus clustering with most variable genes. e Two microenvironment subtypes with different immune/stromal infiltrations among 95 patients were identified by unsupervised clustering using curated signatures of 24 microenvironment cell types. Immune enrichment score (IES), stromal enrichment score (SES) and other cohort information were annotated at the top panel. SFCE Société Française du Cancer de l’Enfant, TARGET Therapeutically Applicable Research to Generate Effective Treatments, TCGA The Cancer Genome Atlas, PSL Pitié-Salpêtrière Hospital, DAWT diffuse anaplastic Wilms tumor, FAWT focal anaplastic Wilms tumor, FHWT favorable histology Wilms tumor, ChRCC chromophobe renal cell carcinoma, ccRCC clear cell renal cell carcinoma, PRCC papillary renal cell carcinoma. Source data are provided as a Source Data file.