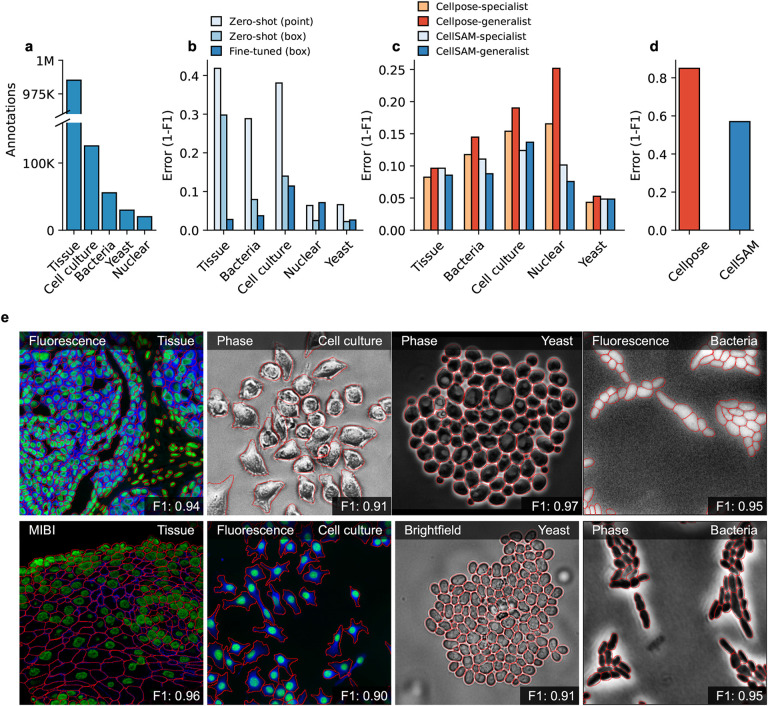
Fig. 2. CellSAM is a strong generalist model for cell segmentation.
a) For training and evaluating CellSAM, we curated a diverse cell segmentation dataset from the literature. The number of annotated cells is given for each data type. Nuclear refers to a heterogeneous dataset (DSB)50 containing nuclear segmentation labels. b) Zero-shot (ZS) and fine-tuned mask generation error (1- F1 score) for SAM when using point and bounding box prompts. All prompting in this figure was done with ground truth prompts. The best performance is achieved with bounding box prompts and fine-tuning. c) Segmentation performance for CellSAM and Cellpose on different data types. We compare the segmentation error (1-F1) for models that were trained as specialists (e.g., on one dataset) or generalists (the full dataset). Models were trained for a similar number of steps across all datasets. We observed that CellSAM-generalhas a lower error than Cellpose-general on almost all tested datasets. Further, we observed that generalist training improved CellSAM’s performance over specialist training; the reverse was true for Cellpose. d) Zero-shot performance of CellSAM-general and Cellpose-General on the LIVECell dataset. Here, we show greater than 4x segmentation performance on an unseen dataset. e) Qualitative results of CellSAM segmentations for different data and imaging modalities. Predicted segmentations are outlined in red.