Extended Data Fig. 7 |. Influence of valine at P7 of VL96B on CD94/NKG2A recognition.
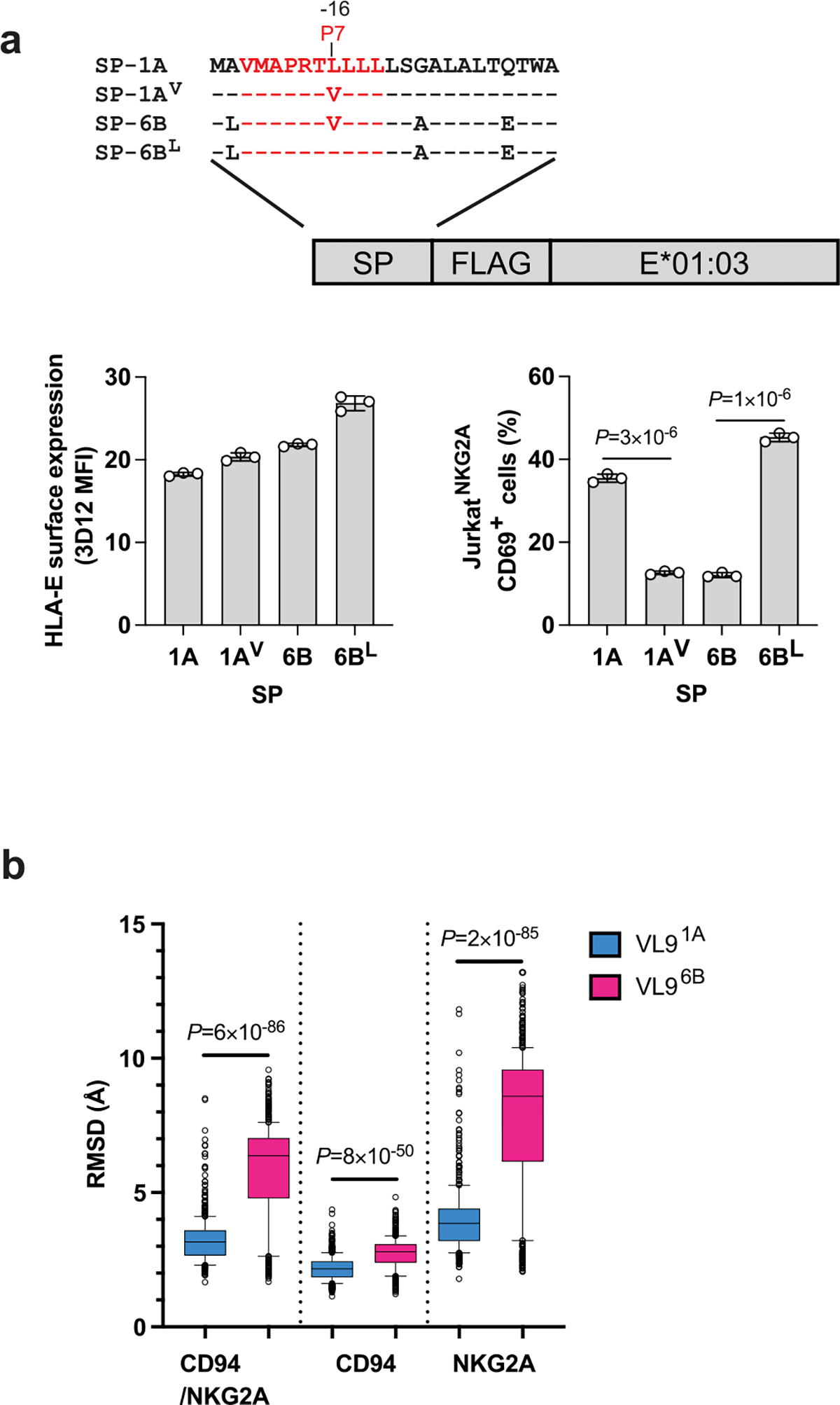
a, Mutation analysis of P7 using JurkatNKG2A reporter cell response to .221-SPE*01:03 cells. Schematic representation of hybrid swap constructs transduced into .221 cells is shown on top. HLA-E expression level on .221-SPE*01:03 cells expressing wild-type (1A, 6B) and mutant (1AV, 6BL) SPs, and JurkatNKG2A activity against these cells are shown below. Data represent triplicate experiments. Error bars represent the mean ± SD. P values for comparisons between wild type and mutant SPs were determined by a two-sided unpaired t-test. b, Molecular dynamics simulation analysis of CD94/NKG2A–HLA-E*01:03 complexes in the presence of VL91A and VL96B peptides. Box plots show the root-mean-square deviations (RMSDs) of the full receptor, CD94, or NKG2A depending on the presence of the two distinct VL9 peptides in the binding groove during a 5 μs simulation (n = 500). Box boundaries span the 25–75 percentiles with the median marked in the middle; the whiskers extend to 10 and 90 percentiles, and the remaining data points are shown as gray circles. The RMSD box plots demonstrate higher receptor motions in VL96B-loaded complexes P values for comparisons between VL91A and VL96B were determined by a two-sided Mann-Whitney test.
