Figure EV2. RNase H activity is dispensable for the activation of early replication origins and the repression of late origins in the presence of HU.
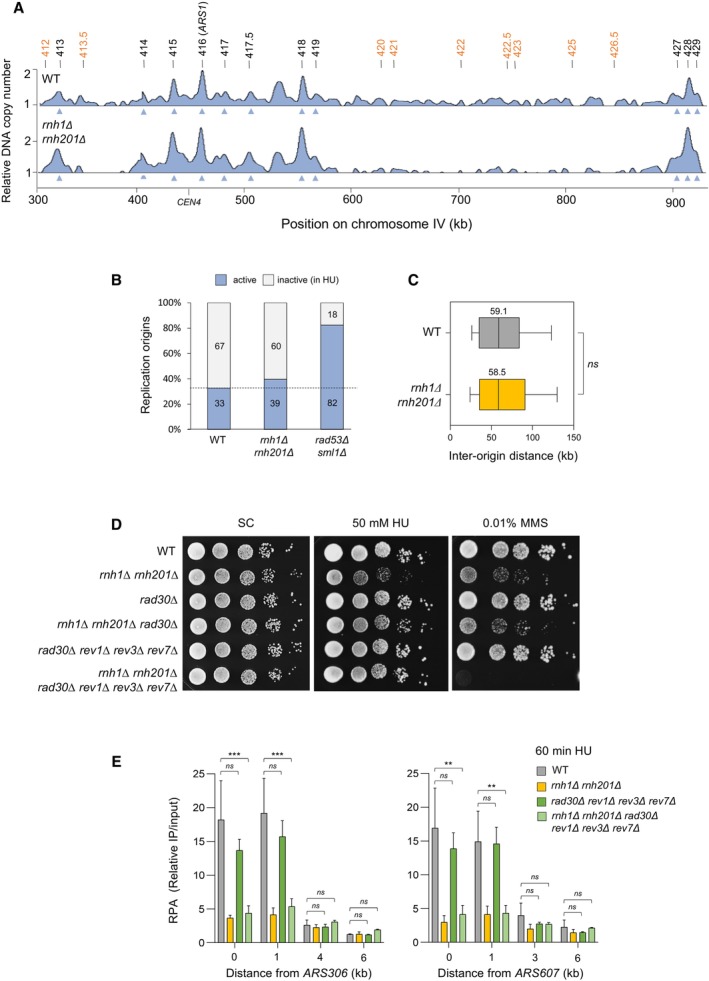
- Genome‐wide analysis of origin usage in wild‐type and rnh1Δ rnh201Δ cells released synchronously into S phase for 60 min in the presence of 200 mM HU. Relative DNA copy number was determined by deep sequencing as the ratio of normalized reads in HU and G1 cells. A representative region on chromosome IV is shown. Positions of early (black) and late (orange) origins are indicated. Arrowheads point to active origins.
- Number of active and inactive origins in the experiment shown in panel (A). The rad53Δ sml1Δ mutant is used as a positive control for the derepression of late origins in HU (Poli et al, 2012).
- Distribution of inter‐origin distances determined by DNA combing after releasing cells from G1 into S phase for 90 min in the presence of 200 mM HU. Box, 25–75 percentile range. Whiskers, 10–90 percentiles range. Median is indicated in kb. ns: not significant, Mann–Whitney rank‐sum test. The DNA combing experiment was repeated twice (n = 2 biological replicates) with similar results, one representative experiment is shown.
- Growth of the indicated strains on synthetic complete (SC) ± 50 mM HU or 0.01% MMS. Spots correspond to 1:10 serial dilutions.
- ChIP‐qPCR analysis of RPA enrichment around ARS306 and ARS607 in the indicated cells released synchronously into S phase for 60 min in the presence of 200 mM HU. RPA enrichment was normalized to four unreplicated regions. Mean and SEM are indicated (n = 3 biological replicates). For statistical analysis, two‐way ANOVA was applied. **P < 0.01; ***P < 0.001.
