Figure 5. RNase H2 promotes the resection of nascent DNA in HU‐treated HeLa cells.
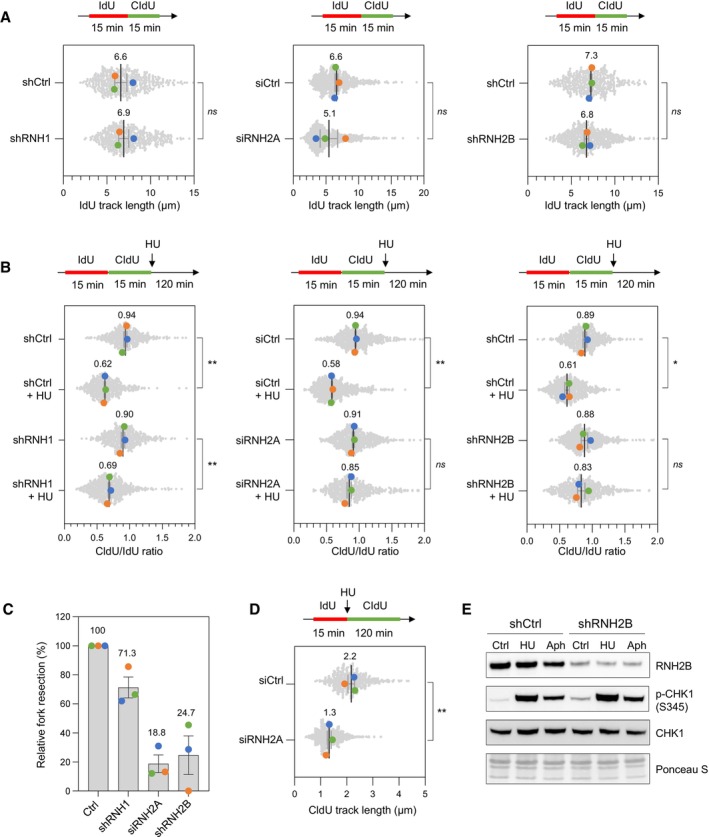
- DNA fiber analysis of replication fork speed in control HeLa cells and in cells depleted for RNase H1 (shRNH1), RNase H2A (siRNH2A) or RNase H2B (shRNH2B). Control (shCtrl), shRNH1 and shRNH2B cells were treated with doxycycline (10 μg/ml) for 72 h. Control (siCtrl) and siRNH2A cells were transfected with siRNAs for 48 h. Cells were sequentially labeled with IdU and CldU for 15 min before DNA fiber spreading. The length of IdU tracks from individual forks (~100–300 forks/condition) is shown. Mean (μm, n = 3 biological replicates) and SEM are indicated in gray. ns, non‐significant, paired t test.
- Stalled fork resection was analyzed using DNA fiber spreading. Depletion of RNase H1, RNase H2A or RNase H2B in HeLa cells was performed as indicated in panel (A). After sequential labelling of IdU and CldU for 15 min, cells were either collected immediately or treated for 2 h with 4 mM hydroxyurea (HU) before DNA fiber analysis. The lengths of the IdU and CldU tracks (~100–300 tracks/condition) were plotted as the ratio of CldU to IdU. Mean (n = 3 biological replicates) and SEM are indicated in gray. *P < 0.05; **P < 0.01; ns, non‐significant, paired t test.
- Relative extent of fork resection in control and RNase H‐deficient as determined by DNA fiber spreading (panel B). Mean ± SEM are indicated (n = 3 biological replicates).
- DNA fiber analysis fork progression in HU‐treated control and RNase H2A‐depleted cells. HeLa cells were transfected with siRNA against RNase H2A (siRNH2A) or a control sequence (siCtrl) for 48 h, and then labeled for 15 min with IdU before 4 mM HU treatment for 2 h. The distribution of CldU tracks (~100–300 tracks/condition) length is shown for three biological replicates. Mean (μm, n = 3) and SEM are indicated in gray. **P < 0.01, paired t test.
- Control (shCtrl) and RNH2B‐depleted (shRNH2B) HeLa cells were treated with doxycycline (10 μg/ml) for 72 h. Cells were then treated with or without HU (4 mM) or aphidicolin (1 μM) for 2 h. Activation of CHK1 was detected by Western blotting using anti‐phophoCHK1 (S345) antibody. Total CHK1 and ponceau staining are used as loading controls.
Source data are available online for this figure.
