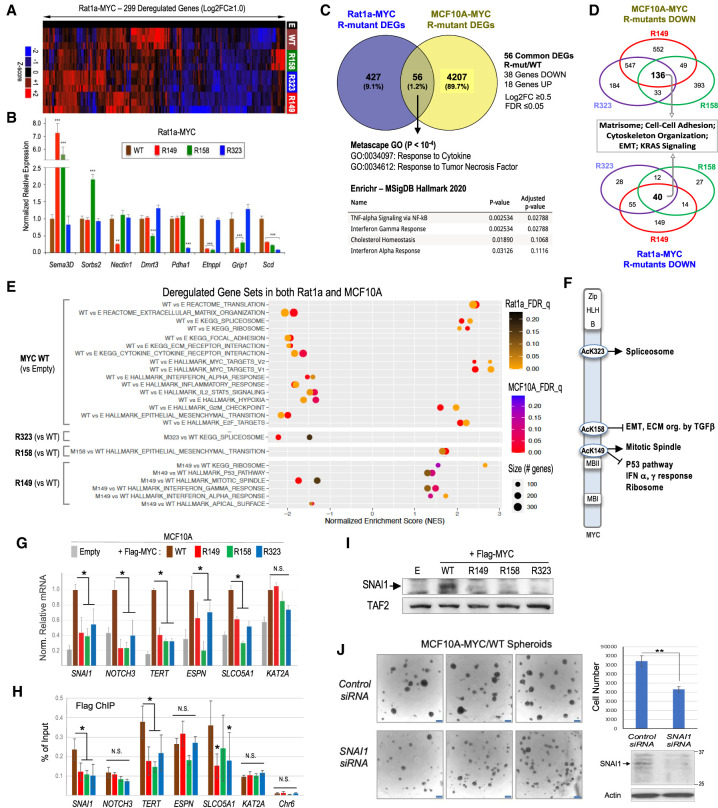
Figure 7.
MYC AcK-dependent genes and pathways in common in Rat1a and MCF10A cells and the role of AcK residues in MYC binding to select target genes. (A) Heat map with unsupervised hierarchical clustering of the top 299 genes deregulated twofold or more (FDR ≤ 0.07) in MYC R mutant Rat1a cells relative to cells overexpressing MYC WT. (B) RT-qPCR analysis of a select group of Rat1a genes deregulated by specific MYC R mutants. Gene mRNA expression was normalized to PPIA. (C) Venn diagram of deregulated genes (DEGs) in common in Rat1a and MCF10A cells (log2FC ≥ 0.5, FDR ≤ 0.05) affected by at least one MYC R mutant relative to MYC WT cells. Gene ontology (GO) terms (Metascape) and most enriched molecular signatures (Enrichr and MSigDB) for the 56 overlapping DEGs are shown. (D) Venn diagrams of genes down-regulated by 1.5-fold or more (FDR ≤ 0.05) in all three MYC R mutant cell lines (relative to MYC WT cells) and associated top GO pathways. (E) GSEA enrichment scores for gene sets deregulated similarly in Rat1a and MCF10A cell lines. (F) Summary of GSEA results shown in E, illustrating the gene sets/pathways enriched (arrows) or inhibited (blunt end arrows) in MYC WT relative to the AcK-to-R mutant cell lines. (G) RT-qPCR analysis of mRNAs of select MYC-activated genes in the indicated MCF10A cell lines. Expression values were normalized to TOP1 and UBC and are relative to expression in the FLAG-MYC WT cell line (set arbitrarily to 1.0). (H) ChIP analyses of FLAG-MYC WT and R mutants binding to the promoters of MYC-activated genes or to an unrelated locus in chromosome 6 (Chr6). (N.S.) No statistical significance (P > 0.05). N ≥ 3 biological replicates each in triplicates (error bars are SD). (I) Western blot analysis of SNAI1 expression in MCF10A-MYC WT and R mutant cell lines. (J) MCF10A-MYC/WT cells were transfected with control or SNAI1 siRNAs and, after 48 h, analyzed for SNAI1 protein expression by Western blot and suspension spheroid growth by microscopy (scale bar, 250 μm) and total spheroid cell counting (bar graph).