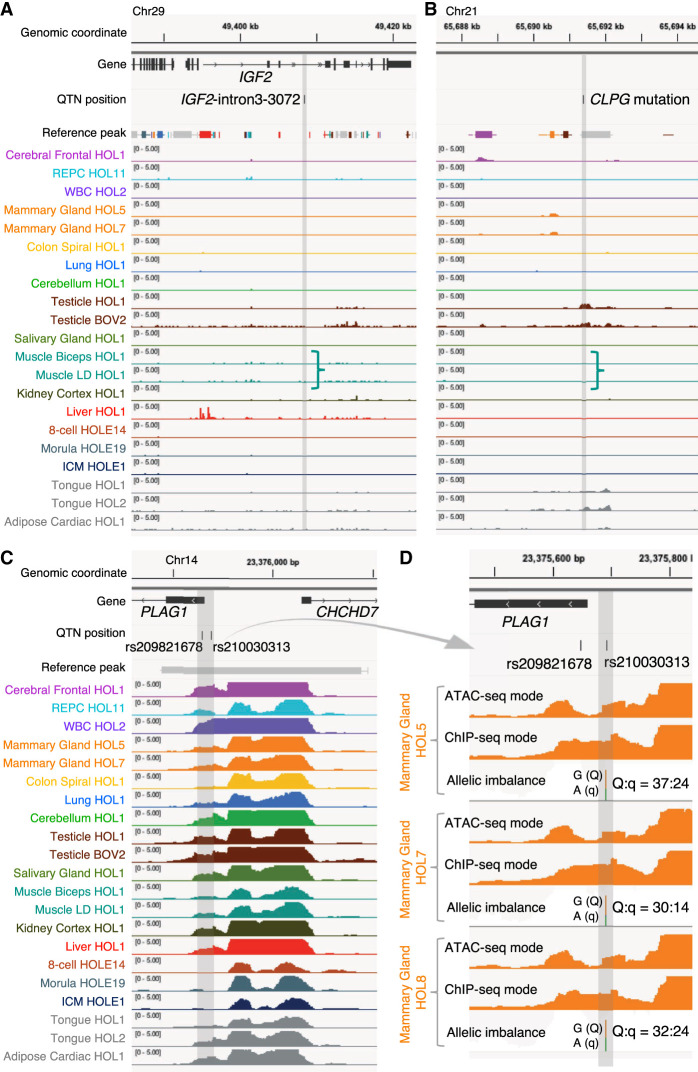
Figure 5.
A retrospective evaluation of the utility of the ATAC-seq catalog for identifying regulatory variants. ATAC-seq peaks at three genomic loci encompassing regulatory QTNs previously identified in domestic animals. Chromosome coordinates, gene annotations, QTN positions, core and consensus reference peak regions (thick bars and horizontal lines, respectively; color-coded based on their highest NFM component), and peaks (ChIP-seq mode tag coverage unless otherwise mentioned) from at least one tissue sample representing each NMF component group with corresponding color code. Positions of the QTNs are highlighted as vertical gray bands. Track height measures the normalized tag coverage (1,000,000/[total tag count]). (A) The bovine orthologous region encompassing the IGF2-intron3-3072 QTN identified in pigs (A/G at susScr11: Chr 2: 1,483,817; bosTau9: Chr 29: 49,408,408) (Van Laere et al. 2003; Markljung et al. 2009) that maps to a 16-bp motif highly conserved among placental mammals disrupts interaction of the ZBED6 repressor, resulting in an approximately threefold up-regulation of IGF2 in postnatal skeletal muscle affecting muscle growth, heart size, and fat deposition. None of ATAC-seq peaks overlapping the 16-bp motif were called across the 104 ATAC-seq data analyzed in this study. (B) The bovine orthologous region encompassing the callipyge (CLPG) muscular hypertrophy mutation identified in sheep (A/G at oviAri4: Chr 18: 64,294,536; bosTau9: Chr 21: 65,691,397) (Freking et al. 2002; Smit et al. 2003). The mutation is located in a 12-bp highly conserved motif among placental mammals and is considered to disrupt a muscle-specific long-range control element (a silencer) that causes ectopic expression of a 327-kb cluster of imprinted genes in postnatal skeletal muscle. ATAC-seq peaks overlapping the mutation site were called only in testis and tongue samples but not in skeletal muscle. (C) Bovine PLAG1 promoter region encompassing two out of eight candidate QTNs influencing bovine stature identified by Karim et al. (2011) (rs209821678 [alternatively ss319607405]: (CCG)11/(CCG)9 at bosTau9: Chr 14: 23375648–23375650; rs210030313 [ss319607406]: G/A at bosTau9: Chr 14: 23375692). The two QTNs reside in a strong 1044-bp-long ubiquitous peak between the PLAG1 and CHCHD7 TSSs. Regions encompassing the other six credible variants do not map to any called peak in our ATAC-seq data and, hence, are not shown. (D) Enlargement of the two QTN loci for three animals that are Qq heterozygous at rs210030313. Peaks called with ATAC-seq and ChIP-seq modes, as well as allelic imbalance in mapped reads, are shown. The two QTNs reside in a footprint of the ATAC-seq mode peak, which is recovered by a ChIP-seq mode peak, indicating the presence of trans-acting factor(s) in the region hindering cleavage events by transposases. Allelic imbalance at rs210030313 (Q = G; q = A) indicates that the Q allele is more accessible compared with the q allele. Previous work showed that the two regulatory variants affect bidirectional promoter strength and that the Q allele, associated with bigger stature, showed approximately 1.5-fold higher promoter activity compared with the q allele in a luciferase assay. Figures were created using the Integrative Genomics Viewer (Robinson et al. 2011).