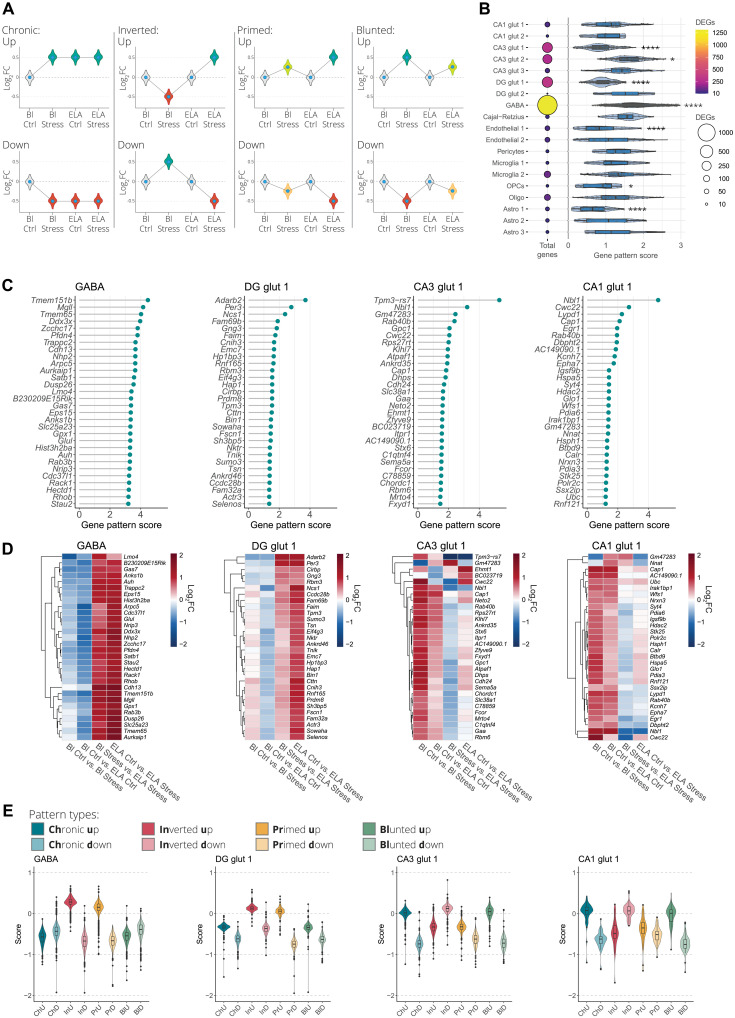
Fig. 3. Widespread and cell type–specific transcriptional patterning induced by ELA.
(A) Illustration of the theoretical transcriptional patterning effects where ELA interacts with gene expression changes induced by acute stress exposure. The transcriptional patterning effects are categorized as up- or down-regulated chronic, inverted, primed, or blunted expression. (B) Dot plot on the left side represents all combined DEGs in 19 clusters identified in all four comparisons (Bl Ctrl versus Bl Stress, Bl Ctrl versus ELA Ctrl, Bl Stress versus ELA Stress, and ELA Ctrl versus ELA Stress). Size and the color of the circles indicate the number of DEGs. The violin plot on the right side indicates the calculated gene patterning score for each DEG associated with each cluster. The gene patterning score of several clusters significantly deviates from the collection of all gene patterning scores from all 19 clusters. The higher the score, the higher the likelihood to find one of the four gene patterning types, while a lower score indicates a lower interaction between ELA and the acute stress. One-way ANOVA with Tukey post hoc test. (C) Lollipop plot ranking the top 30 highest scoring genes in GABAergic neurons (GABA), dentate gyrus (DG) glutamatergic neuron cluster 1 (DG glut 1), CA3 glutamatergic neuron cluster 1 (CA3 glut 1), and CA1 glutamatergic neuron cluster 1 (CA1 glut 1). (D) Clustered heatmap visualization of the log2FC of the top 30 highest scoring genes in the four selected clusters. The four rows in each heatmap represent the four differential expression comparisons. (E) Quantification of each of the individual transcriptional patterning types in the four selected clusters identifies distinct transcriptional patterns. Box plots represent the 25%, median, and 75% quartile; whiskers span 1.5 × IQR. *P < 0.05, ****P < 0.0001.