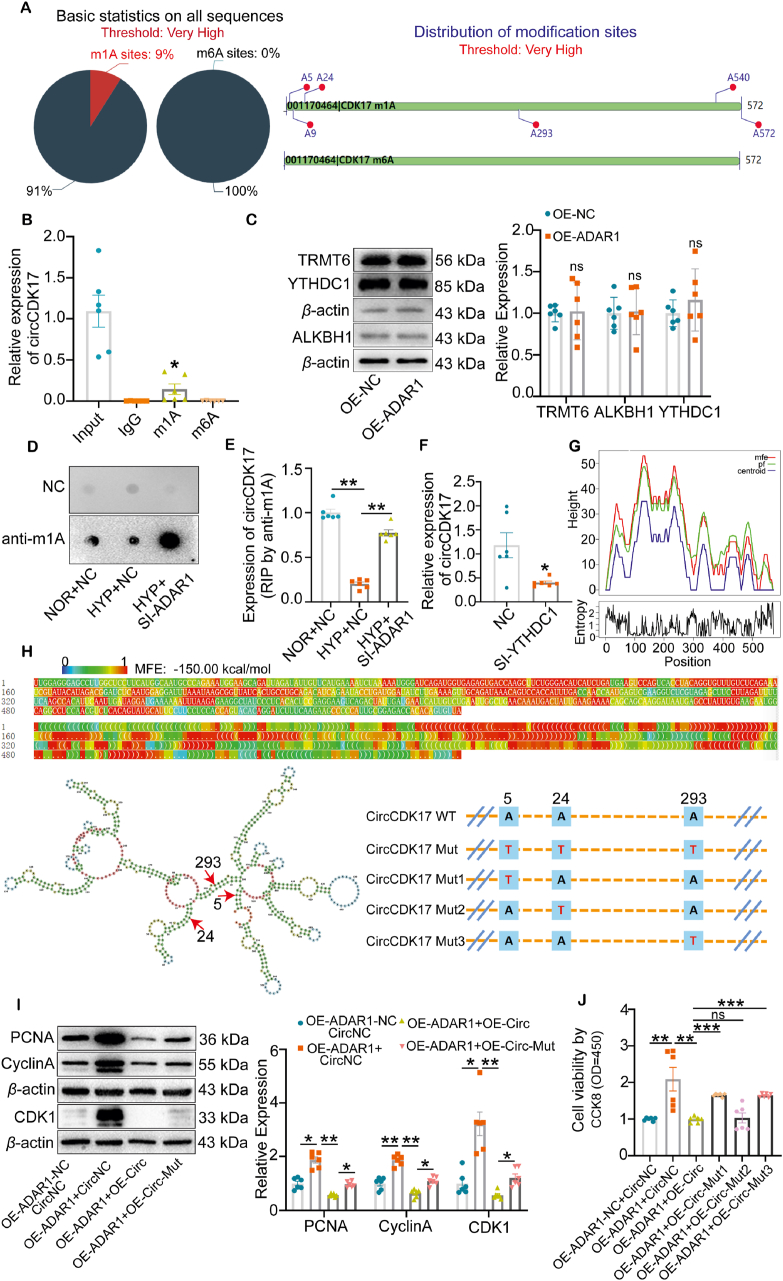
Figure 7.
ADAR1 regulated m1A modification of circCDK17. (A) Online software deeppromise predicts the m1A and m6A modifications of circCDK17 (https://deeppromise.erc.monash.edu/). (B) meRIP-qPCR assay showing the binding of circCDK17 to m1A antibody instead of m6A antibody, n = 6. (C) Western blotting analysis showing protein expression of m1A modifying enzymes in response to ADAR1 overexpression, n = 6. (D) Dot blots showing the modification of m1A upon ADAR1 knockdown. (E) meRIP-qPCR analysis showing the effect of m1A antibody on circCDK17 enrichment in response to the knockdown of ADAR1, n = 6. (F) qPCR analysis showing the expression of circCDK17 upon knockdown of m1A reader YTHDC1, n = 6. (G) Representation of Mountain plot of the MFE (minimum free energy) structure, the partition function (pf), the thermodynamic ensemble of RNA structures, the centroid structure and present the positional entropy for each position (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi). (H) The optimal secondary structure in dot-bracket notation with a minimum free energy of −150.00 kcal/mol is given. RNAfold was combined with the online software catRAPID (http://service.tartaglialab.com/page/catrapid_group) to analyze mutant plasmid sites. (I) Western blotting analysis showing that mutation of all three m1A modification sites attenuated the effects of overexpression of ADAR1 on protein expression of PCNA, Cyclin A, and CDK1, n = 6. (J) CCK8 analysis showing the effect of site mutations of each of the three m1A modification sites on cell viability of PASMCs, n = 6. NOR, normoxia; HYP, hypoxia; NC, negative control; OE, overexpression; Mut, mutant; SI, siRNA; Statistical analysis was performed with two-way ANOVA followed by Dunnett's test or the Student's t-test; All values are presented as mean ± SEM. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.