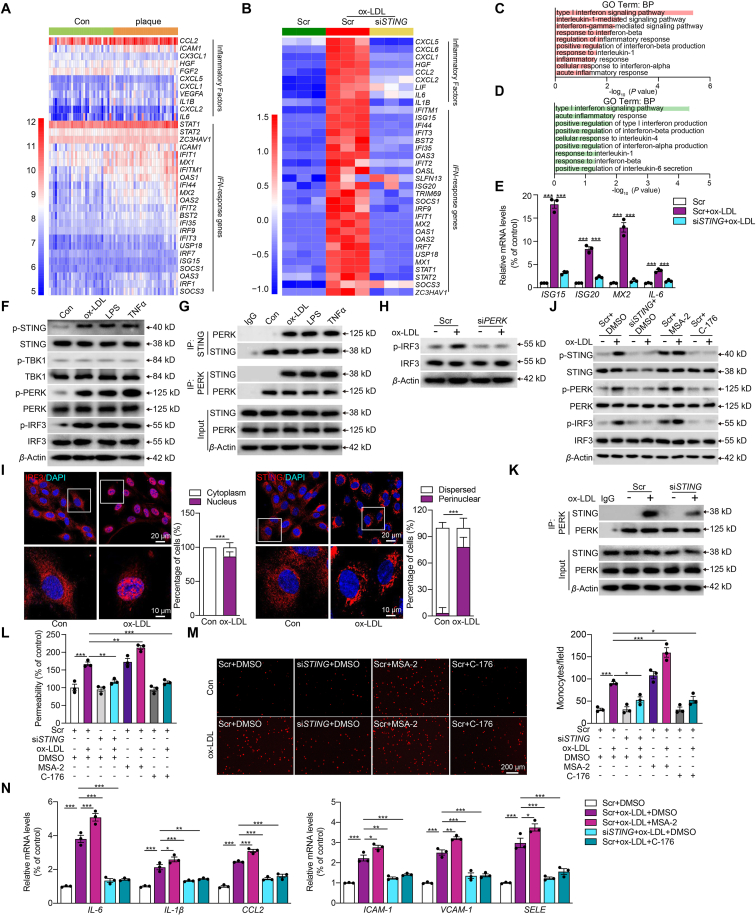
Figure 1.
Atherosclerosis-related endothelial dysfunction mediated by oxidized LDL is dependent on the activation of the non-canonical STING–PERK pathway. (A, B) Heat map displaying the expression (microarray; log2 RMA signal) of inflammatory factors and IFN-response genes in distant macroscopically intact tissue as control (Con, light green) and atherosclerotic plaque (orange) in (A) and human coronary artery endothelial cells (HCAECs) treated with scramble control (Scr, green), Scr + oxidative low-density lipoprotein (ox-LDL, red), and stimulator of interferon genes siRNA (siSTING)+ox-LDL (yellow) in (B). (C, D) GO biological process enrichment in HCAECs treated with Scr + ox-LDL versus Scr (C) and in HCAECs treated with siSTING + ox-LDL versus Scr + ox-LDL (D) by DAVID analysis. (E) Quantitative real-time PCR (qRT-PCR) analysis of ISG15, ISG20, MX2, and IL-6 mRNA expression in ox-LDL-treated HCAECs combined with siSTING. (F) Western blot analysis of the non-canonical STING–PKR-like ER kinase (PERK) pathway in HCAECs treated with ox-LDL (100 μg/mL), lipopolysaccharide (LPS, 1 μg/mL), or tumor necrosis factor α (TNFα, 10 ng/mL). (G) Co-immunoprecipitation analysis of STING-PERK binding in HCAECs treated with ox-LDL, LPS and TNFα. (H) Western blot analysis of the phosphorylated interferon regulatory factor 3 (p-IRF3) in ox-LDL-treated HCAECs combined with siPERK. (I) Immunofluorescence staining of STING and IRF3 in ox-LDL-treated HCAECs versus control (Scale bar = 20 μm). (J) Western blot analysis of the non-canonical STING–PERK pathway in ox-LDL-treated HCAECs combined with siSTING, STING inhibitor C-176 (0.5 μmol/L) or STING activator MSA-2 (10 μmol/L). (K) Co-immunoprecipitation analysis of STING–PERK binding in ox-LDL-treated HCAECs combined with siSTING. (L, M) Permeability of HCAECs to FITC-Dexstran (L), THP-1 monocytes adhesion (M) in ox-LDL-treated HCAECs combined with siSTING or MSA-2 or C-176. (N) qRT-PCR analysis of adhesions molecules and chemokines mRNA expression in ox-LDL-treated HCAECs combined with siSTING or MSA-2 or C-176. Data are expressed as mean ± SEM, n = 3; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.