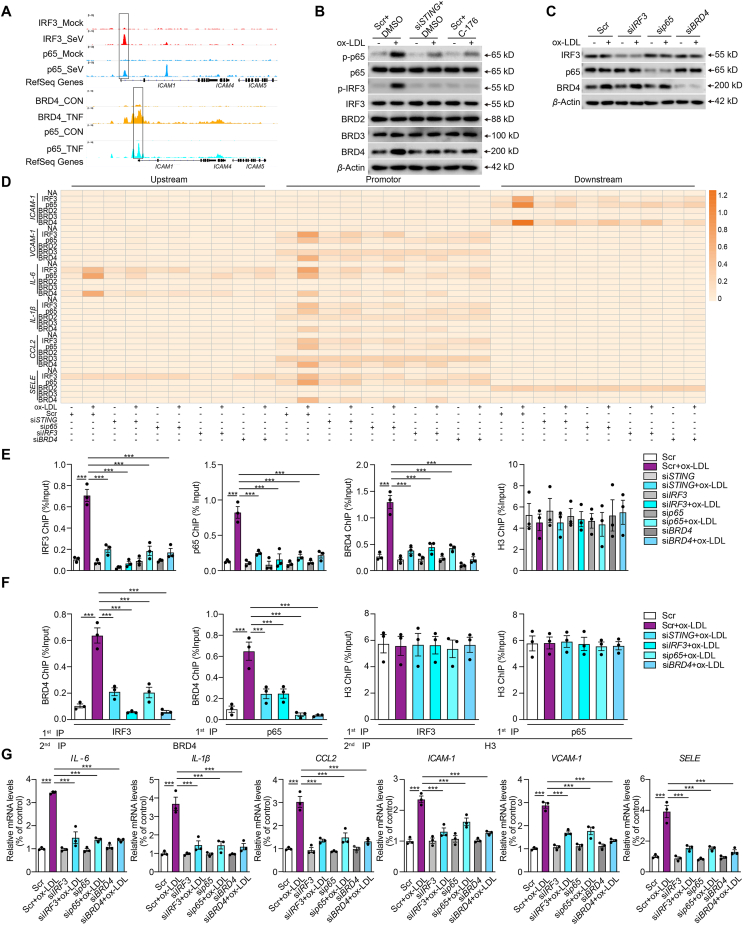
Figure 4.
Cooperation of IRF3, NF-κB and BRD4 in the transcription of inflammation genes in oxidized LDL-induced endothelial injury. (A) Tracks of chromatin immunoprecipitation sequencing (ChIP-seq) peaks for IRF3 (red), p65 (blue, upper; cyan, down), and BRD4 (yellow) at the ICAM1 gene loci in control (top) or stimulant-induced inflammation (bottom) samples. (B) Western blot analysis of total and phosphorylated p65 and IRF3 and BRD family in ox-LDL-treated HCAECs combined with siSTING or C-176. (C) Western blot analysis of total and phosphorylated p65 and IRF3 and BRD4 in ox-LDL-treated HCAECs combined with IRF3, p65, or BRD4 siRNA. (D) ChIP analysis of the enrichment of IRF3, p65, BRD2, BRD3, and BRD4 at −1 kb, promoter and +1 kb of inflammatory genes in ox-LDL-treated HACECs combined with STING, p65, IRF3 or BRD4 siRNA. (E, F) The ChIP-qPCR (E) and Re-ChIP (F) analysis of the binding of BRD4, IRF3, p65, and IRF3 on the +1 kb region of ICAM1 in ox-LDL-treated HCAECs combined with STING, p65, IRF3 or BRD4 siRNA. (G) qRT-PCR analysis of adhesions molecules and chemokines mRNA expression in ox-LDL-induced HCAECs combined with STING, p65, IRF3 or BRD4 siRNA. Data are shown as mean ± SEM, n = 3; ∗∗∗P < 0.001.