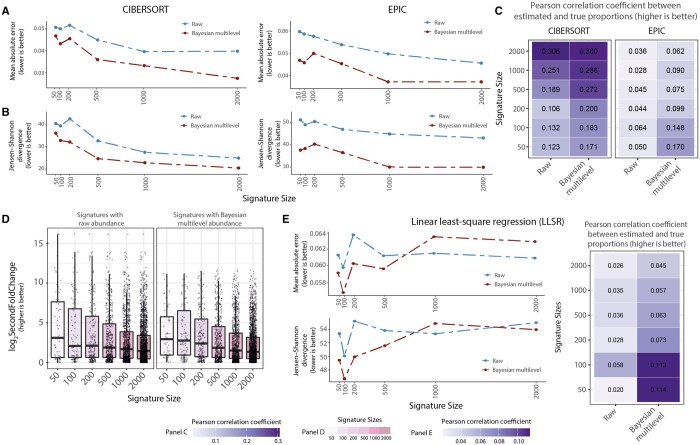
Figure 5.
cellsig improves the CIBERSORTx estimation of cell-type transcriptomic signatures with multilevel transcriptional adjustment. (A) Benchmark of CIBERSORTx-generated transcriptional signatures with varying sizes from raw or adjusted transcript abundance. The deconvolution accuracy (for CIBERSORTx and EPIC) was measured with the median (across 100 in silico mixtures) mean-absolute-error values. (B) The deconvolution accuracy was measured with Jensen–Shannon divergence. (C) Pearson’s correlation coefficient values between the ground truth and estimated proportions from the deconvolutions. (D) Boxplots of second-fold-change scoring of the signatures generated by CIBERSORTx. The second-fold-change scores the difference in abundance of the markers between cell types where they are expressed highest and second highest. Boxplots are arranged and colour-coded for signature sizes. (E) Deconvolution benchmark using llsr from signatures generated by CIBERSORTx. (F) Pearson's correlation coefficient between the ground truth and estimated proportions from the llsr deconvolution method.