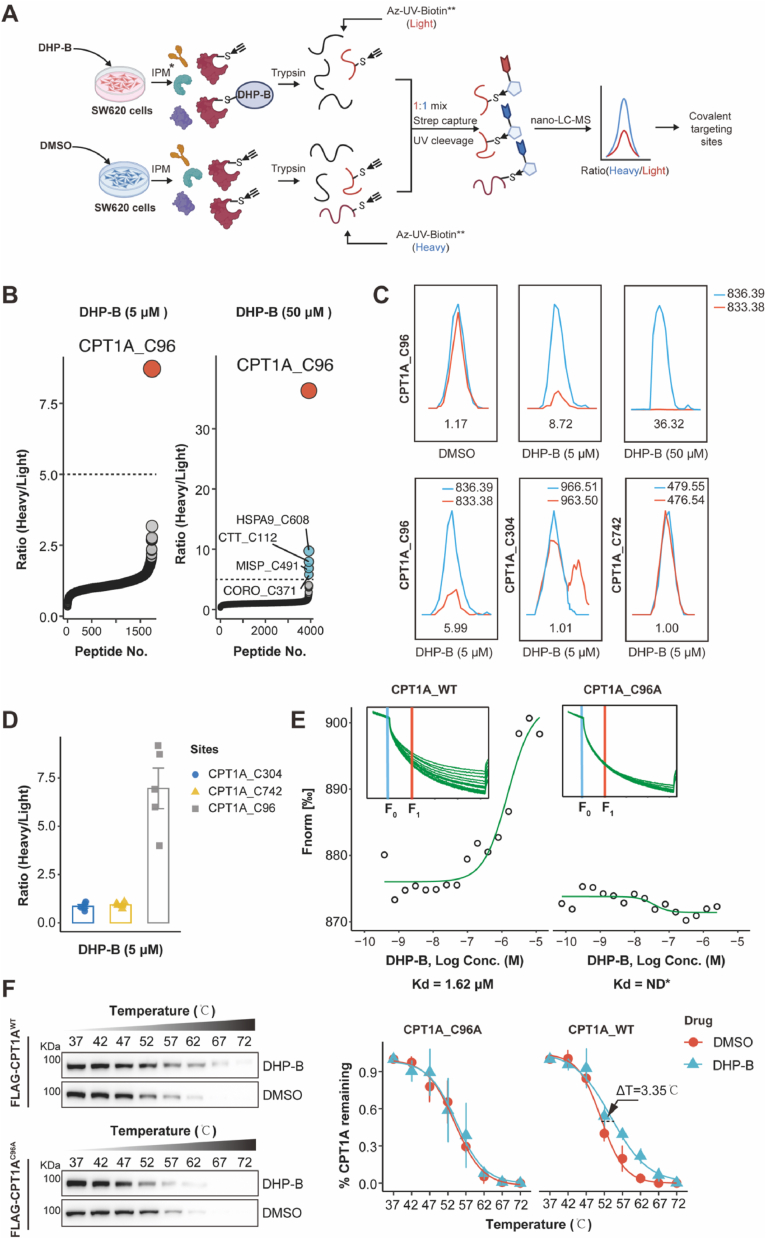
Fig. 4.
Screening and molecular biological validation of covalent targets of DHP-B against CRC. (A) The proteome from DHP-B or DMSO-treated SW620 cells was tagged with an alkynyl 3-indoleacetic acid (IPM) probe in vitro and digested by trypsin. The peptides were conjugated with Light (L) and Heavy (H) UV cleavable biotin-azide (Az-UV-biotin) reagents, combined and collected using streptavidin beads. Photo-releasing probe-modified peptides for LC-MS/MS analysis and pFind3 post-processing. (B) Distribution of competitive QTRP ratios (RatioH/L) from SW620 reactions with 5 μM or 50 μM of DHP-B (n = 5). A threshold of five-fold or more IPM probe labeling blockage (RatioH/L > 5) marks cysteines that are sensitive to DHP-B, and proteins with the most competitive DHP-B reactivity are named. (C) Representative MS1 profiles for DHP-B-sensitive C96 peptide in CPT1A showing concentration-dependent IPM labeling blockade (n = 5). (D) MS1 spectrum and RatioH/L for active cysteine-containing peptides from the CPT1A protein; among the three peptides only C96 demonstrates activity to 5 μM DHP-B competition (n = 5). The RatioH/L data mean ± SD is provided. (E) MST dose-response curves of GFP-labeled CPT1AWT or CPT1AC96A mutant bound to DHP-B. Dissociation constants (Kd) values were also exhibited (n = 5). (F) CETSA showed the thermal stability of CPT1AWT or CPT1AC96A mutant proteins following treatment with DHP-B (5 μM). Right panel: CETSA curve and the thermal stability to reach 50 % of temperature (Tm) value was performed using GraphPad Prism software, n = 5, presented as Mean ± SD.