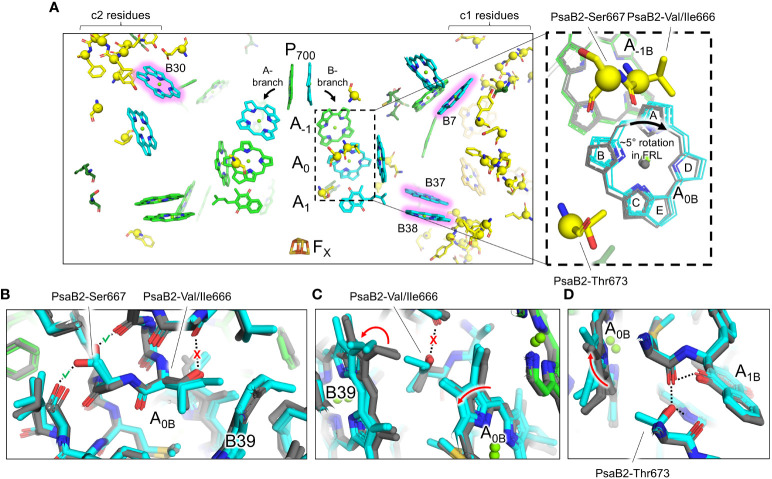
Figure 6.
FRL-specific residues near the ETC cofactors and rotation of the Chl molecule in site A0B. (A) Structural view of FRL-PSI from F. thermalis PCC 7521 (PDB 7LX0) centered on the ETC cofactors. Chl molecules are shown as tetrapyrrole rings only. Chl f sites are shown with a pink glow. Phylloquinones are shown as the headgroup with a truncated tail. FRL-specific residues are shown as sticks with a sphere for their Cα atom, the latter of which is consistent with spheres shown in Figure 3 . The dashed box region is magnified in the right panel. Here, the residues shown are from the structure of FRL-PSI from F. thermalis PCC 7521, but additional cofactors are shown from the following structures: FRL-PSI from Synechococcus sp. PCC 7335 (PDB 7S3D, colored), FRL-PSI from H. hongdechloris (PDB 6KMX, colored), VL-PSI from H. hongdechloris (PDB 6KMW, grey), non-FaRLiP PSI from T. elongatus (PDB 1JB0, grey), and non-FaRLiP PSI from Synechocystis sp. PCC 6803 (PDB 5OY0, grey). For the A0B cofactor, the tetrapyrrole rings are labeled A-E. (B–D) Views of the protein near the A0B cofactor and FRL-specific alterations that cause its rotation. The structures of FRL-PSI (colored) from Synechococcus sp. PCC 7335 and H. hongdechloris, and VL-PSI (grey) from T. vulcanus and H. hongdechloris are shown. Dashed lines denote H-bonding interactions. Where dashed lines are covered by a red “X”, the H-bond is unique to VL-PSI structures. Where dashed lines are covered by a green check mark, the H-bond is unique to FRL-PSI structures. Red arrows indicate where rotations occur in FRL-PSI structures relative to VL-PSI structures.