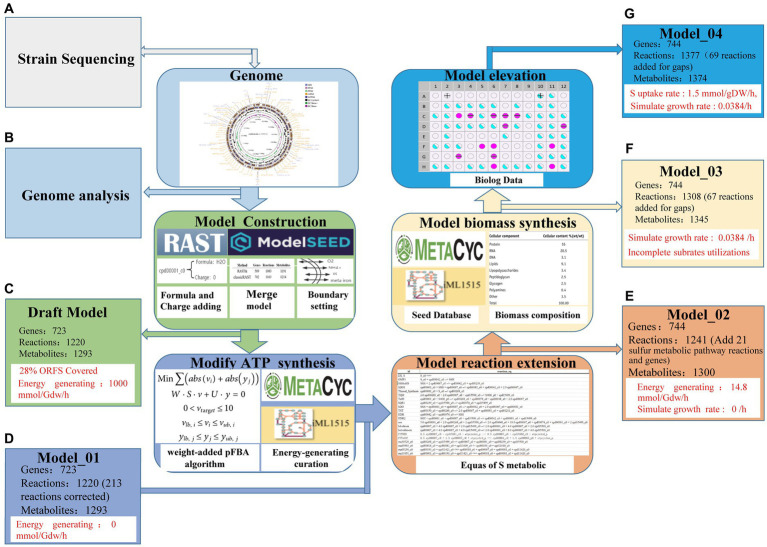
Figure 4.
The reconstruction process of genome-scale metabolic network workflow for Acidithiobacillus AMEEHan. (A) Whole genome sequencing of the strain, perform whole genome sequencing of samples to obtain gene sequence data. (B) Analysis of bacterial whole genome, analyze genome sequences to obtain gene function annotation information. (C) Draft model generation and brief information, predict initial draft metabolic network model containing some gaps based on genome annotation. (D) Model_01 generation from Draft Model, refine and correct wrong reactions in Draft model using knowledge bases like Metacyc to produce Model_01. (E) Model_02 generation from Model_01, further integrate literature knowledge and add some known biochemical reaction pathways to generate Model_02. (F) Model_03 generation from Model_02, perform gap-filling on the Model_02 using the pFBA algorithm to produce Model_03. (G) Final model Model_04, based on Biolog experimental results, expand Model_03 to generate the final high-quality model Model_04.