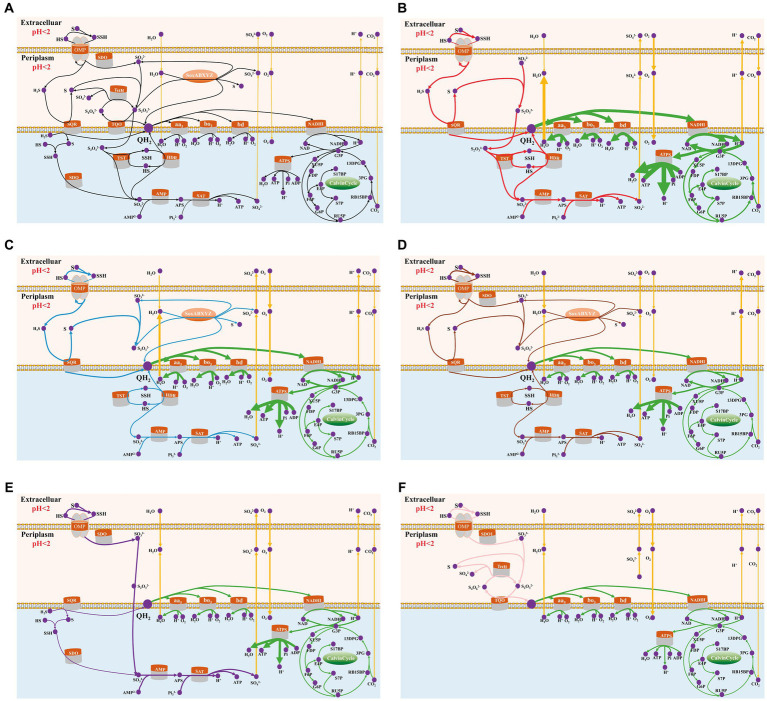
Figure 7.
Sulfur oxidation pathway of Acidithiobacillus AMEEHan in silico predictions. The green line showed the common sulfur oxidation reactions found in all pathways while the yellow lines indicated intracellular and extracellular transport of substances. The purple dots represented the corresponding metabolites. Arrows indicate the direction of enzymatic activity and the arrow thicknesses are proportional to the flux through each reaction (a thicker arrow has a larger flux). (A) The potential sulfur oxidation pathways in Acidithiobacillus AMEEHan. (B) The red line denoted the optimal pathway predicted for sulfur oxidation. (C) The blue line denoted the second pathway predicted for sulfur oxidation. (D) The brown line denoted the third pathway predicted for sulfur oxidation. (E) The purple line denoted the fourth pathway predicted for sulfur oxidation. (F) The pink line denoted the fifth pathway predicted for sulfur oxidation. S, Sulfur; SSH, Sulfane sulfur atom of glutathione persulfide; OMP, Outer-membrane proteins; TQO, Thiosulfate quinone oxidoreductase; TetH, Tetrathionate hydrolase; SoxABXYZ, Sulfur oxidizing enzyme system; SQR, Sulfide: quinone oxidoreductase; SDO, Sulfur dioxygenase; TST, Rhodanese; HDR, HDR-like complex; SAT, ATP sulfurylase; aa3, bd, bo3, terminal oxidases; QH2, Quinol pool; NADHI, NADH dehydrogenase complex I; and ATPs, ATP hydrolytic enzyme.