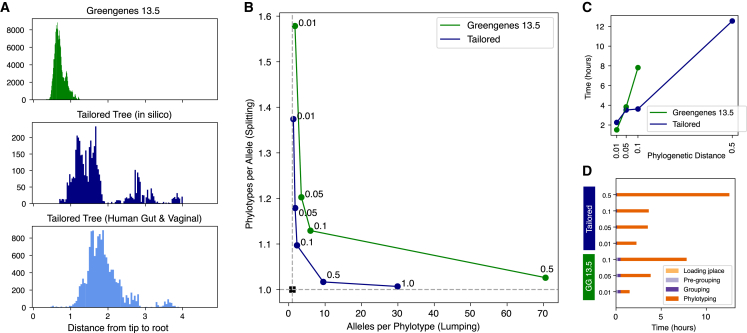
Figure 2.
A comparison of phylotypes from the off-the-shelf Greengenes versus custom-tailored de novo phylogenetic tree reveals superior performance of the tailored de novo tree
(A) Tip-to-root distances for the Greengenes 13.5 tree (top), and de novo phylogenetic trees generated for ASVs from the in silico (middle) or collection of human vaginal and gut microbiota (bottom), with the de novo trees comprising tips of full-length non-clustered 16S rRNA SSU alleles. Depicted as a histogram, with the x axis the tip-to-root distance and the y axis the count of tips in that bin of distances.
(B) Scatterplot of the mean 16S rRNA SSU gene alleles-per-phylotype (x axis) versus phylotypes-per-allele (y axis) of phylotypes generated after placement on the Greengenes 13.5 phylogeny (green) or de novo “tailored” phylogenetic tree (blue) at various binning distances (as labeled next to data points). The ideal outcome is marked as a black square, at one allele per phylotype and one phylotype per allele.
(C) Relationship between computational wall time in hours and phylogenetic distance of binning on 87,477 ASVs from six different studies of the human gut microbiome and three of the human vaginal microbiome after placement on two different trees (a tailored tree via MaLiAmPi in navy blue, same as in A; Greengenes 13.5 in green). Testing was conducted on an AMD 5900X CPU, Python 3.10.12 and with Taichi version 1.6.0, llvm 15.0.4. Phylotyping of the Greengenes 13.5 tree at a distance of 0.5 took over 48 h and is not depicted.
(D) Computational wall time in hours broken down by the four major steps for phylotyping: loading the placement file, pre-grouping, grouping, and phylotyping. This was for the handling of 87,477 ASVs from six different studies of the human gut microbiome and three of the human vaginal microbiome after placement on two different trees (a tailored tree via MaLiAmPi, same as in A; Greengenes 13.5) at phylogenetic binning distances of 0.01, 0.05, 0.1, and 0.5. Testing was conducted on an AMD 5900X CPU, Python 3.10.12 and with Taichi version 1.6.0, llvm 15.0.4. Phylotyping of the Greengenes 13.5 tree at a distance of 0.5 took over 48 h and is not depicted.