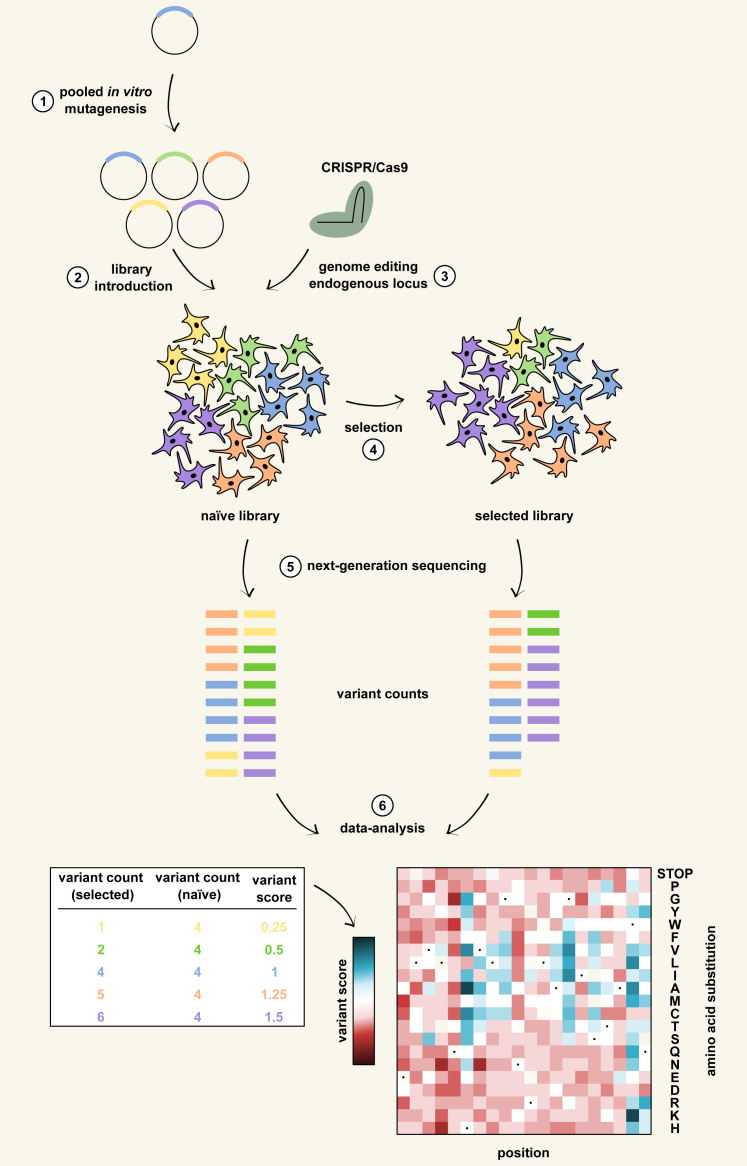
Figure 1.
The deep mutational scanning workflow
A schematic overview of the workflow for deep mutational scanning of target proteins in mammalian cells. The different steps follow the sections within the text. (1) Library generation through pooled in vitro mutagenesis. (2) Library introduction as a single variant per cell. (3) Library generation through large-scale genome editing of the endogenous locus. CRISPR-Cas9-based systems generate one variant in each cell. (4) Library screening through functional selection. Functional assays quantitatively couple the function of a variant to a selectable phenotype so that selective pressure changes the frequency of the variant in the library in accordance with its function. Selectable phenotypes are typically a fitness effect for growth selection or a fluorescent signal for fluorescence-activated cell sorting (FACS). (5) Quantification of variant function by sequencing during selection. Next-generation sequencing (NGS) allows the quantification of the selection-induced frequency change of each variant in the library. (6) Data analysis. The quantitative information from NGS is used to calculate a functional score for each variant, typically visualized on a heatmap.