Figure 2.
Deep immunological imprinting caused by the ancestral spike in bivalent COVID-19 mRNA vaccines impaired serum-neutralizing antibody responses to Omicron subvariants
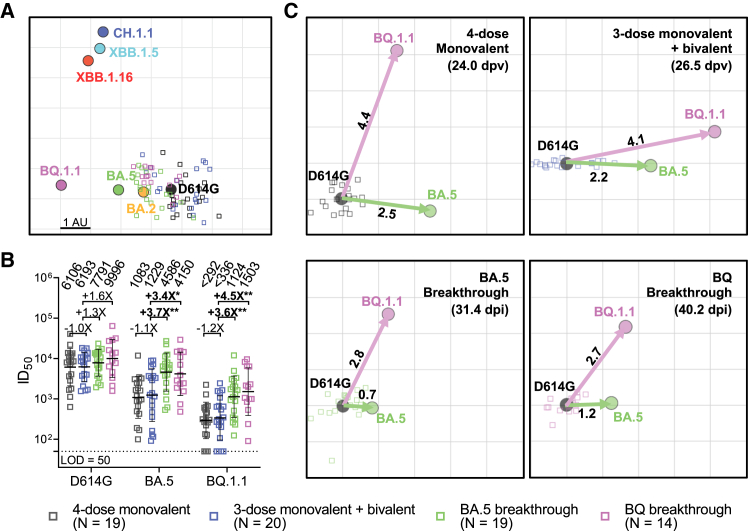
(A) Antigenic map derived from the neutralization data in Figure 1B. SARS-CoV-2 variants and sera are shown as colored circles and squares, respectively. The x and y axes represent antigenic units (AU), with each grid corresponding to a 2-fold serum dilution of the neutralization titer, as defined by the RACMACS package (https://github.com/acorg/Racmacs/tree/master).
(B) Neutralizing antibody responses induced by a fourth dose of a bivalent mRNA vaccine compared to a fourth dose of the original monovalent booster or breakthrough infections. The values above the symbols indicate the geometric mean ID50 titer for each cohort. The assay limit of detection (LOD = 50) is represented by a dotted line. Comparisons were made against 3-dose monovalent + bivalent cohort by Mann-Whitney test, and the statistical significance is represented as ∗p < 0.05 or ∗∗p < 0.01. The numbers in parentheses represent the mean days post last vaccination (dpv) or the mean days post infection (dpi).
(C) Antigenic maps for individual cohorts against D614G, BA.5, and BQ.1.1. Arrows indicate the antigenic distances from D614G to BA.5 (green) and BQ.1.1 (magenta). See also Figure S1.