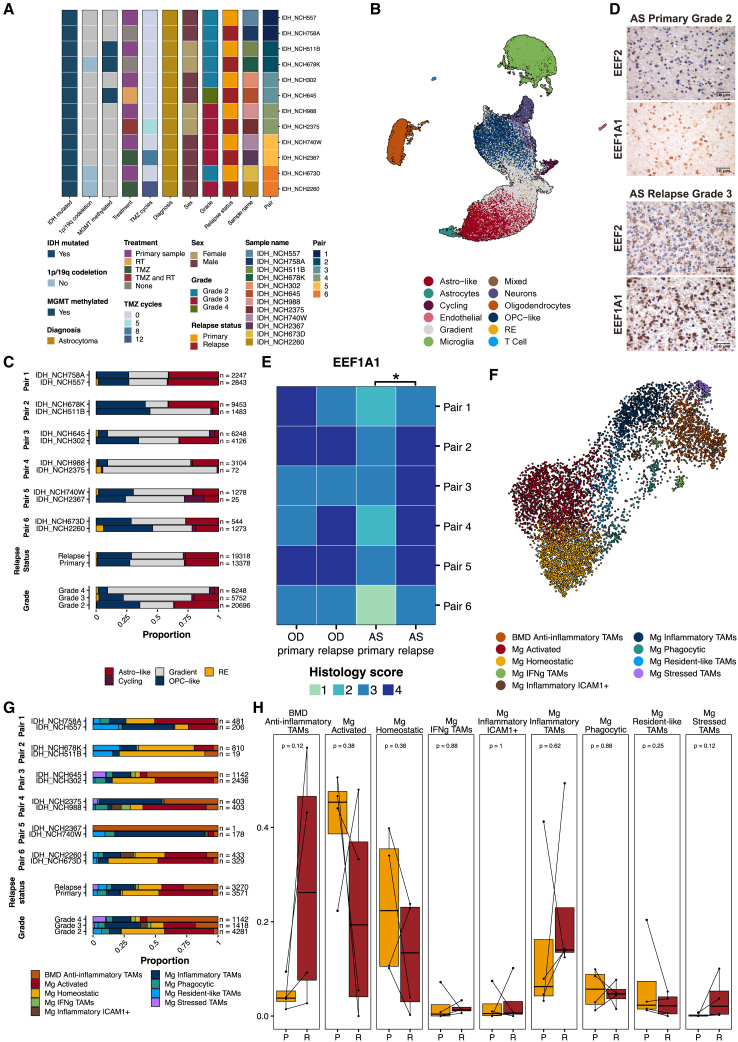
Figure 4.
snRNA-seq of paired primary and recurrent AS cohorts indicates shifts in tumor populations associated with grade
(A) Clinical and molecular characteristics of 6 paired primary and recurrent IDH-mutant AS cohorts for snRNA-seq.
(B) UMAP embedding of integrated snRNA-seq data from paired AS tumors, colored by assigned cell types. UMAP1, x axis; UMAP2, y axis.
(C) Bar plots showing tumor population proportions, separated by pairs, relapse status, and grade.
(D) Representative IHC staining for RE markers EEF2 and EEF1A1 in paired primary (grade 2) and relapse (grade 3) AS tumors. Images are captured at 40x original magnification (scale bar, 50 μm)
(E) Heatmap showing semi-quantitative EEF1A1 histological scores (0 = 0%, 1 = 0%–5%, 2 = 6%–29%, 3 = 30%–69%, 4 >70% positive staining) for 6 pairs of primary and recurrent OD and AS tumors. Wilcoxon signed-rank test for paired samples was performed to test for significance, ∗p < 0.05.
(F) UMAP embedding of integrated microglia population of paired primary and recurrent AS cohorts, colored by assigned TAM subpopulations. UMAP1, x axis; UMAP2, y axis.
(G) Bar plots showing TAM subpopulation proportions separated by pairs, relapse status, and grade.
(H) Boxplots showing TAM subpopulation proportions separated by relapse status. P, primary; R, relapse.