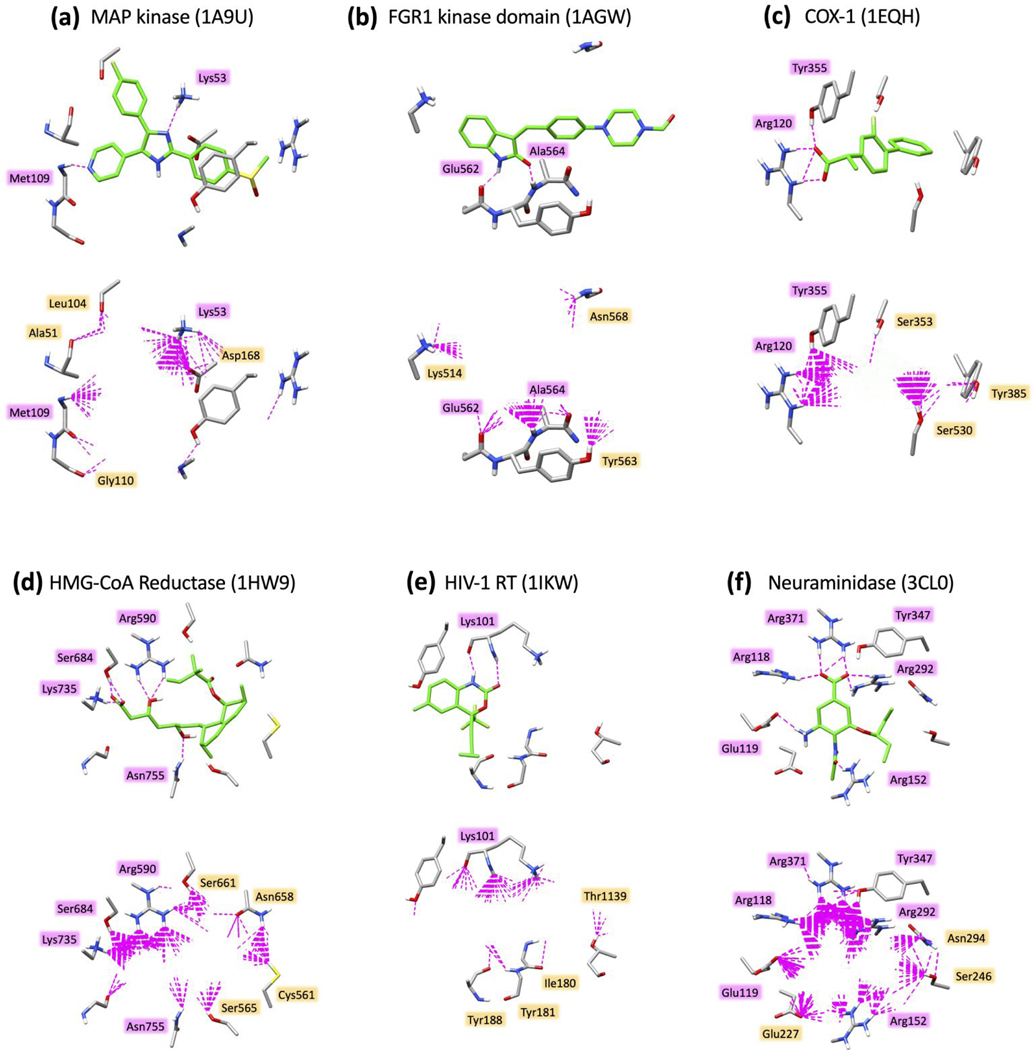
Figure 17.
H-bond patterns (dashed magenta lines) made by six reference ligands (top panels, ref ligand pose in green) and top-scoring ensembles (N=500 molecules) from D3N-pinpoint simulations (bottom panels, D3N molecules hidden for clarity) for (a) SB203580 with Map kinase (1A9U), SU2 with FGR1 kinase domain (1AGW), flurbiprofen with COX-1 (1EQH), simvastatin HMG-CoA reductase (1HW9), efavirenz with HIV-1RT (1IKW), and oseltamivir with neuraminidase (3CL0). H-bonds calculated using the Chimera59 program with default settings. Select binding site residues shown in gray.