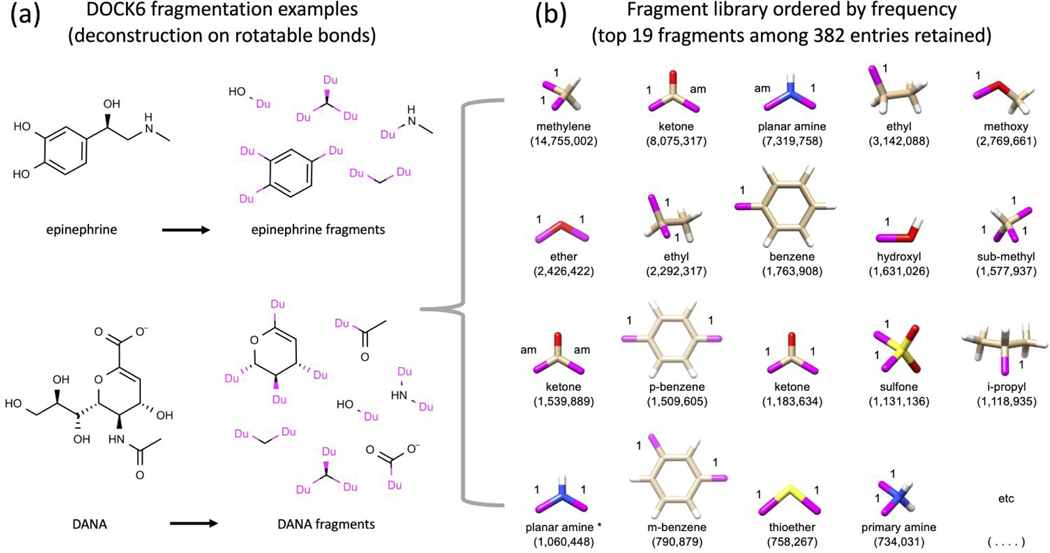
Figure 2.
Schematic illustrating (a) how DOCK6 fragment libraries are derived by deconstructing molecules along rotatable bonds and (b) the top 19 fragments ordered by frequency (out of 382 retained) for a library derived from 13M drug-like molecules downloaded from ZINC.2–4 Based on the number of dummy atoms (magenta attachment points) the fragments are classified into sidechains (1 attachment point, N=217), linkers (2 attachment points, N=146), or scaffolds (3+ attachment points, N=19).