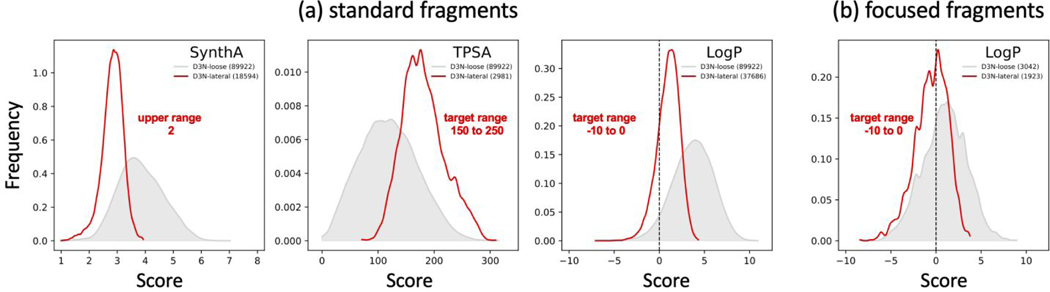
Figure 7.
Normalized descriptor populations using (a) the standard DOCK6 fragment library or (b) a focused fragment library derived from molecules with negative LogP values. In each case, a single descriptor with laterally shifted target ranges (D3N-lateral, red) were used to drive de novo growth in the absence of protein. Results obtained using the D3N-loose protocols (gray) are shown as a control. Legends indicate the specific target ranges (labeled in red font) and the number of molecules obtained (in parenthesis). Readers should note this data was derived from four independent experiments. The standard deviations employed for D3N-lateral protocols are the same as listed in Table 3 for D3N-drugc. TPSA results in angstroms squared.