Abstract
We previously reported that hepatitis C virus (HCV) infection activates the reactive oxygen species (ROS)/c-Jun N-terminal kinase (JNK) signaling pathway. Activation of JNK contributes to the development of liver diseases, including metabolic disorders, steatosis, liver cirrhosis and hepatocellular carcinoma. JNK is known to have numerous target genes, including JunB, a member of activator protein-1 transcription factor family. However, the roles of JunB in the HCV life cycle and HCV-associated pathogenesis remain unclear. To clarify a physiological role of JunB in HCV infection, we investigated the phosphorylation of JunB in HCV J6/JFH1-infected Huh-7.5 cells. Immunoblot analysis revealed that HCV-induced ROS/JNK activation promoted phosphorylation of JunB. The small interfering RNA (siRNA) knockdown of JunB significantly increased the amount of intracellular HCV RNA as well as the intracellular and extracellular HCV infectivity titers. Conversely, overexpression of JunB significantly reduced the amount of intracellular HCV RNA and the intracellular and extracellular HCV infectivity titers. These results suggest that JunB plays a role in inhibiting HCV propagation. Additionally, HCV-mediated JunB activation promoted hepcidin promoter activity and hepcidin mRNA levels, a key factor in modulating iron homeostasis, suggesting that JunB is involved in HCV-induced transcriptional upregulation of hepcidin. Taken together, we propose that the HCV-induced ROS/JNK/JunB signaling pathway plays roles in inhibiting HCV replication and contributing to HCV-mediated iron metabolism disorder.
Keywords: Hepatitis C virus, ROS/JNK/JunB, Viral propagation
INTRODUCTION
According to the World Health Organization, an estimated 58 million people worldwide are chronically infected with hepatitis C virus (HCV), and 1.5 million new infections occur each year, highlighting that HCV remains a significant public health concern (1). HCV is a leading cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (2). Despite the fact that direct-acting antivirals (DAAs) therapy has a high sustained virological response rate of over 95% in patients with various HCV genotypes, the emergence of DAAs resistance and limited access to DAAs therapy in developing countries continue to pose challenges to global efforts to eliminate HCV (3, 4).
HCV is a positive-sense single-stranded RNA virus that belongs to the Hepacivirus genus of the Flaviviridae family. The HCV genome consists of 9.6-kb RNA encoding a single polyprotein of about 3,010 amino acids, which is processed by viral proteases and cellular signalases to produce three structural proteins (Core, E1, and E2) and seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (5).
We previously reported that HCV infection induces reactive oxygen species (ROS) production and activates the c-Jun N-terminal kinase (JNK) signaling pathway, leading to the enhancement of hepatic gluconeogenesis (6) and the induction of apoptosis (7). Recently, we demonstrated that the HCV-mediated ROS/JNK signaling pathway promotes the release of HCV particles via activation of the E3 ubiquitin ligase Itch (8). The activated JNK pathway regulates various physiological processes, such as cell death, differentiation, proliferation, and carcinogenesis through phosphorylation of numerous targets, including JunB, a member of the activator protein-1 (AP-1) transcription factor family (9–11). Transcription factor JunB has been implicated in controlling proliferation, apoptosis, and malignant transformation (12, 13) by regulating transcription of target genes, such as cyclin-dependent kinase inhibitor p16 (INK4a) (14) and matrix metalloproteinase 2 (15). However, the roles of JunB in the HCV life cycle and HCV-associated pathogenesis are largely unknown.
Hepatic iron overload has been shown to be one of the pathological features in patients with HCV-related chronic liver diseases (16, 17). Several lines of evidence indicate that hepatic iron overload is associated with hepatocarcinogenesis in chronic hepatitis C (18). However, the mechanisms underlying HCV-mediated hepatic iron accumulation have not been fully elucidated. Hepcidin, a liver-derived peptide hormone, plays a major role in maintaining iron homeostasis. Increased hepcidin expression leads to elevated hepatic iron storage (19, 20). Hepcidin expression in the hepatocytes is primarily regulated at the transcriptional level. Notably, the JNK/JunB signaling pathway is involved in iron metabolism via the activation of hepcidin transcription (21).
In this study, we demonstrate that the HCV-induced ROS/JNK signaling pathway activates the transcription factor JunB. We demonstrate evidence suggesting that JunB plays roles in inhibiting HCV replication and upregulation of hepcidin. Thus, we propose that the ROS/JNK/JunB signaling pathway negatively regulates HCV replication and also contributes to iron metabolism disorder via the activation of hepcidin transcription.
MATERIALS AND METHODS
Cell culture and virus
A human hepatoma cell line, Huh-7.5 cells (22) was kindly provided by C. M. Rice (The Rockefeller University, NY). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (High Glucose) with L-glutamine (FUJIFILM Wako Pure Chemical Industries, Osaka, Japan) and supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin (Gibco, Grand Island, NY), 0.1 mM nonessential amino acids (Gibco), and 10% heat-inactivated fetal bovine serum (Biowest, Nuaillé, France). Cells were transfected with plasmid DNA using FuGene 6 transfection reagents (Promega, Madison, WI).
The pFL-J6/JFH1 plasmid, which encodes the entire viral genome of a chimeric strain of HCV-2a, JFH1 (23), was kindly provided by C. M. Rice. The HCV genome RNA was synthesized in vitro using pFL-J6/JFH1 as a template and was transfected into Huh-7.5 cells by electroporation. The virus produced in the culture supernatant was used for the titration of virus infectivity (24). Virus infection was performed at a multiplicity of infection (MOI) of 1 in the infection experiments. Huh-7 cells stably harboring an HCV-1b full-genome replicon (FGR) derived from Con1 (RCYM1) were also used (25).
Plasmids
A full-length JunB cDNA was amplified by reverse transcription-PCR (RT-PCR) using mRNAs of Huh-7.5 cells as a template. The primer sequences were as follows: sense primer, 5′-TACCGAGCTCGGATCATGTGCACTAAAATGGAACAG-3′; antisense primer, 5′-GATATCTGCAGAATTCGAAGGCGTGTCCCTTGAC-3′. The amplified PCR product was purified and inserted into the BamH1/EcoR1 sites of pEF1A-Myc-His6 (Invitrogen, Carlsbad, CA) using an In-Fusion HD cloning kit (Clontech, Mountain View, CA).
Antibodies and reagents
The mouse monoclonal antibodies (MAbs) used in this study were anti-HCV core MAb (24) (clone 2H9; a kind gift from T. Wakita, National Institute of Infectious Diseases, Tokyo, Japan), anti-NS3 MAb (MAB8691, Millipore, Billerica, MA), anti-c-Myc MAb (9E10; sc-40, Santa Cruz Biotechnology, CA), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) MAb (014-25524; FUJIFILM Wako Pure Chemical Industries). The rabbit monoclonal antibodies (RMAbs) used in this study were anti-JunB (C37F9) RMAb (3753S, Cell Signaling Technology, Beverly, MA), anti-phospho-JunB (Thr102/Thr104) (D3C6) RMAb (8053S, Cell Signaling Technology), anti-c-Jun (60A8) RMAb (9165S, Cell Signaling Technology), and anti-phospho-c-Jun (Ser63) RMAb (9261S, Cell Signaling Technology). The rabbit polyclonal antibody (PAb) used in this study was anti-HCV NS5A PAb (clone 2914-1; a kind gift from T. Wakita, National Institute of Infectious Diseases, Tokyo, Japan). Horseradish peroxidase (HRP)-conjugated anti-mouse IgG (7076; Cell Signaling Technology) and HRP-conjugated anti-rabbit IgG (7074; Cell Signaling Technology) were used as secondary antibodies.
HCV NS5A inhibitor daclatasvir (ChemScene, Monmouth Junction, NJ), antioxidant N-acetyl cystein (NAC) (Sigma, St. Louis, MO), and JNK inhibitor SP600125 (Selleck Chemicals, Houston, TX, USA) were used.
Immunoblot analysis
Immunoblot analysis was performed as described previously (7). The cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Millipore). The membranes were incubated with a primary antibody, followed by incubation with an HRP-conjugated secondary antibody. The positive bands were visualized using enhanced chemiluminescence western blotting detection reagents (ECL; GE Healthcare, Buckinghamshire, UK).
siRNA transfection
HCV-infected Huh-7.5 cells or HCV FGR RCYM1 cells were transfected with 40 nM of JunB siRNA (SI00034706; Qiagen, Valencia, CA) using Lipofectamine RNAiMAX transfection reagent (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Allstars negative control siRNA (Qiagen) was used as a negative control. All siRNAs were transfected into cells once.
RNA extraction and real-time RT-PCR
Total cellular RNA was isolated using a ReliaPrep RNA cell miniprep system (Promega) according to the manufacturer’s instructions, and cDNA was generated by using a GoScript reverse transcription system (Promega). Real-time RT-PCR was performed using TB Green Premix Ex Taq II (TaKaRa Bio, Shiga, Japan) with SYBR green chemistry on a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). The primer sequences were as follows: Hepcidin, 5′-ACCAGAGCAAAGCTCAAGACC-3′ and 5′-CAGGGCAGGTAGGTTCTAACG-3′; p16, 5′-GAGCAGCATGGAGCCTTC-3′ and 5′-CCTCCGACCGTAACTATTCG-3′. As an internal control, human GAPDH gene expression levels were measured using the primers 5′-GCCATCAATGACCCCTTCATT-3′ and 5′-TCTCGCTCCTGGAAGATGG-3′.
Luciferase reporter assay
The human hepcidin promoter reporter plasmid, pGL4.10-hepcidin promoter, was kindly provided by T. Suzuki (Department of Virology and Parasitology, Hamamatsu University School of Medicine, Hamamatsu, Japan). HCV-infected Huh-7.5 cells or the uninfected control cells cultured in a 24-well tissue culture plate were transiently transfected with pGL4.10-hepcidin promoter together with pRL-CMV-Renilla (Promega), which expresses Renilla luciferase, was used as an internal control. Luciferase reporter assay was performed using a dual-luciferase reporter assay system (Promega). Firefly and Renilla luciferase activities were measured with a GloMax 96 microplate luminometer (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity for each sample.
Titration of HCV infectivity
Titration of extracellular or intracellular HCV infectivity was performed as described previously (8, 26). The extracellular HCV was collected from cell culture supernatants, and centrifuged at 3,000 × g for 10 min at 4°C to remove cell debris. The intracellular HCV was collected by freeze-thaw cycles of cell lysates. Briefly, HCV-infected cells were washed with phosphate-buffered saline (PBS), harvested by trypsin treatment, and centrifuged at 1,000 × g for 5 min at 4°C. To release intracellular HCV particles, the cell pellets were resuspended in complete DMEM and subjected to five freeze-thaw cycles using liquid nitrogen and a 37°C water bath. Cell debris was removed by centrifugation at 20,400 × g for 20 min at 4°C.
Samples were then used to inoculate Huh-7.5 cells in triplicate. The cells were incubated with the inoculum for 3 h at 37°C and then supplemented with fresh complete DMEM. At 24 h post-inoculation, the cells were fixed in cold 100% methanol for 20 min at −20°C. The fixed cells were subjected to indirect immunofluorescence staining to detect HCV core protein using anti-HCV core MAb (clone 2H9). Alexa Fluor 488-conjugated goat anti-mouse IgG (A11001; Molecular Probes, Eugene, OR) was used for the secondary antibody. The viral titer is expressed as focus-forming units per milliliter of the sample (FFU/mL), determined by the average number of HCV core-positive foci.
Quantification of intracellular HCV RNA
To quantitate intracellular HCV RNA, the total cellular RNA was isolated using a ReliaPrep RNA Cell Miniprep System (Promega) according to the manufacturer’s instructions. The cDNA was generated using a GoScript Reverse Transcription System (Promega). Real-time RT-PCR was performed using TB Green Premix Ex Taq II (Tli RNaseH Plus) (TaKaRa Bio) with SYBR green chemistry on the StepOnePlus Real-Time PCR system (Applied Biosystems, Foster City, CA) as reported previously (8). The primer sequences were as follows: HCV, 5′-AGACGTATTGAGGTCCATGC-3′ and 5′-CCGCAGCGACGGTGCTGATAG-3′. As an internal control, human GAPDH gene expression levels were measured using the primers 5′-GCCATCAATGACCCCTTCATT-3′ and 5′-TCTCGCTCCTGGAAGATGG-3′.
Statistical analysis
Results were expressed as means ± standard errors of the means (SEM). Statistical significance was evaluated by analysis of variance (ANOVA) and was defined as a P value of <0.05.
RESULTS
HCV infection promotes phosphorylation of JunB at Thr102/Thr104 via ROS/JNK signaling pathway
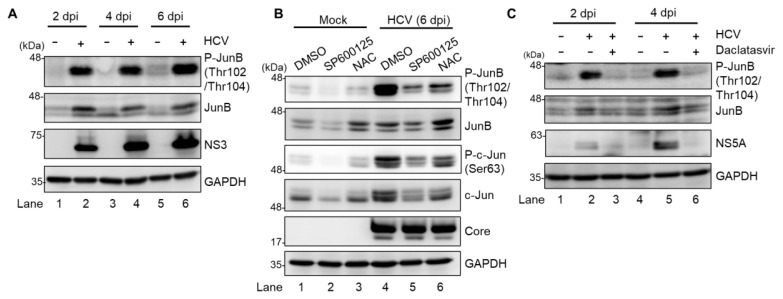
To identify a novel role of the ROS/JNK signaling pathway in the HCV life cycle, we first performed immunoblot analysis to determine whether HCV infection induces phosphorylation and activation of JunB in Huh-7.5 cells. Immunoblot analysis showed that the amount of activated JunB phosphorylated at Thr102/Thr104 markedly increased in HCV J6/JFH1-infected Huh-7.5 cells at 2, 4, and 6 days postinfection (dpi) compared to those in the mock-infected control cells (Fig. 1A, 1st panel, lanes 2, 4, and 6). Notably, the total expression levels of JunB in HCV-infected cells were higher than those in the mock-infected control cells (Fig. 1A, 2nd panel, lanes 2, 4, and 6), suggesting that phosphorylation of JunB stabilizes JunB in HCV-infected cells.
Fig. 1. HCV infection promotes phosphorylation of JunB at Thr102/Thr104 via ROS/JNK signaling pathway.
(A) Huh-7.5 cells were infected with HCV J6/JFH1 at a multiplicity of infection (MOI) of 1. At 2, 4, and 6 days postinfection (dpi), the cells were harvested. The cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (B) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 6 dpi, the cells were harvested with or without pretreatment with either JNK inhibitor SP600125 (30 μM for 30 h) or antioxidant N-acetyl cysteine (NAC; 5 mM for 8 h). The cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (C) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1 with and without treatment of HCV NS5A inhibitor (daclatasvir; 1 μM). At 2 and 4 dpi, the cells were harvested, and the cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control.
To determine whether ROS/JNK signaling pathway induces JunB activation, we treated HCV-infected cells with either the specific JNK inhibitor SP600125 or the antioxidant NAC. We performed immunoblot analysis to determine the phosphorylation status of JunB in HCV-infected cells at 6 dpi. The HCV-induced JunB phosphorylation at Thr102/Thr104 was markedly reduced after treatment with SP600125 or NAC (Fig. 1B, 1st panel, lanes 4–6). In parallel, treatment with SP600125 or NAC clearly decreased the phosphorylation of c-Jun and the total c-Jun protein levels, a key substrate for JNK (Fig. 1B, 3rd and 4th panels, lanes 4–6). These results suggest that HCV infection promotes phosphorylation and activation of JunB via the ROS/JNK signaling pathway.
To further verify the involvement of HCV in the phosphorylation of JunB, we treated the cells with daclatasvir, an HCV NS5A inhibitor, to eliminate HCV. Immunoblot analysis showed that treatment of the cells with daclatasvir decreased HCV NS5A protein (Fig. 1C, 3rd panel, lanes 2 to 3 and 5 to 6) at 2 and 4 dpi. Treatment of the cells with daclatasvir also reduced the HCV-induced phosphorylation of JunB at Thr102/Thr104 (Fig. 1C, 1st panel, lanes 2 to 3 and 5 to 6). These results indicate that HCV infection promotes phosphorylation of JunB.
JunB suppresses HCV replication in HCV J6/JFH1-infected Huh-7.5 cells
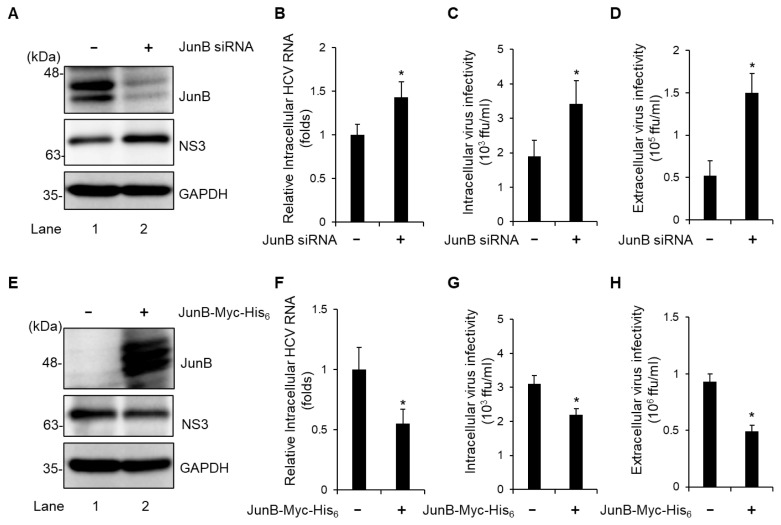
To determine the effect of JunB on the HCV life cycle, Huh-7.5 cells were infected with HCV J6/JFH1 in the presence or absence of JunB small interfering RNA (siRNA). At 3 dpi, we examined the amount of intracellular viral protein, HCV RNA, and HCV infectivity titers as well as extracellular HCV infectivity titers. Endogenous JunB protein was markedly reduced by transfection of JunB siRNA into HCV-infected cells (Fig. 2A, 1st panel, lane 2). Knockdown of JunB increased the level of intracellular HCV NS3 protein (Fig. 2A, 2nd panel, lane 2), the amount of intracellular HCV RNA (Fig. 2B), intracellular HCV infectivity titers (Fig. 2C), and extracellular HCV infectivity titers (Fig. 2D). Conversely, overexpression of JunB reduced the level of intracellular HCV NS3 protein (Fig. 2E, 2nd panel, lane 2), the amount of intracellular HCV RNA (Fig. 2F), intracellular HCV infectivity titers (Fig. 2G), and extracellular HCV infectivity titers (Fig. 2H). These results suggest that JunB suppresses HCV replication in HCV J6/JFH1-infected Huh-7.5 cells.
Fig. 2. JunB suppresses HCV replication in HCV J6/JFH1-infected Huh-7.5 cells.
(A) Huh-7.5 cells were transfected with 40 nM JunB siRNA or control siRNA. At 24 h post-transfection, cells were infected with HCV J6/JFH1 at an MOI of 1. The cells were harvested at 3 dpi. The cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (B) Total cellular RNA was extracted, and HCV RNA was quantitated by real-time RT-PCR. Amounts of intracellular HCV RNA were normalized to amounts of GAPDH mRNA. (C) The cell pellets were washed and disrupted by freeze-thaw cycles, and then the intracellular infectivity titer was measured by a focus-forming assay. (D) The extracellular virus infectivity in the culture supernatants was measured by a focus-forming assay. (E) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with pEF1A-JunB-Myc-His6 or pEF1A-Myc-His6. At 72 h after plasmid transfection, the cells were harvested and cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (F) The total cellular RNA was extracted, and HCV RNA was quantitated by real-time RT-PCR. Amounts of intracellular HCV RNA were normalized to amounts of GAPDH mRNA. (G) The cell pellets were washed and disrupted by freeze-thaw cycles, and then the intracellular infectivity titer was measured by a focus-forming assay. (H) The extracellular infectivity titer in the culture supernatants was measured by a focus-forming assay. Data represent means ± SEM of data from three independent experiments. *, P < 0.05, compared with the controls.
JunB suppresses HCV replication in HCV full-genome replicon (FGR) Con1 (RCYM1) cells
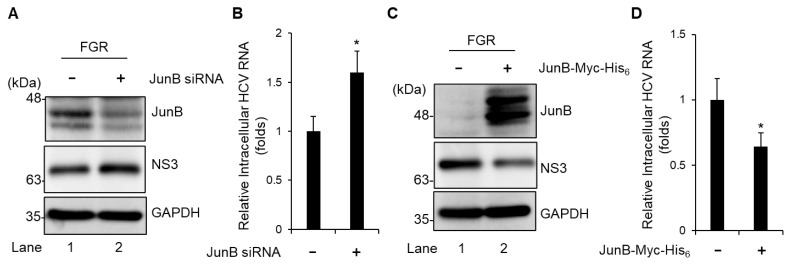
To further assess the effect of JunB on HCV genotype 1b replication, we used Huh-7 cells stably harboring an HCV-1b FGR derived from Con1 (RCYM1). Transfection of JunB siRNA efficiently knocked down endogenous JunB protein in HCV FGR-harboring cells (Fig. 3A, 1st panel, lane 2). Knockdown of JunB increased the level of intracellular HCV NS3 protein (Fig. 3A, 2nd panel, lane 2) and the amount of intracellular HCV RNA (Fig. 3B). Conversely, Overexpression of JunB protein reduced the level of intracellular HCV NS3 protein (Fig. 3C, 2nd panel, lane 2) and significantly decreased the amount of intracellular HCV RNA (Fig. 3D). These results suggest that JunB suppresses HCV replication in both HCV genotypes 2a and 1b.
Fig. 3. JunB suppresses HCV replication in HCV FGR Con1 (RCYM1) cells.
(A) Huh-7 cells stably harboring the HCV-1b FGR RNA derived from Con1 (RCYM1) were transfected with 40 nM JunB siRNA or control siRNA. At 48 h post-transfection, the cells were harvested. The cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (B) Total cellular RNA was extracted and HCV RNA was quantitated by real-time RT-PCR. Amounts of intracellular HCV RNA were normalized to amounts of GAPDH mRNA. (C) HCV FGR Con1 (RCYM1) cells were transfected with either pEF1A-JunB-Myc-His6 or pEF1A-Myc-His6. At 48 h post-transfection, the cells were harvested. The cell lysates were analyzed by immunoblotting with the indicated antibodies. The level of GAPDH served as a loading control. (D) Total cellular RNA was extracted and HCV RNA was quantitated by real-time RT-PCR. Amounts of intracellular HCV RNA were normalized to amounts of GAPDH mRNA. Data represent means ± SEM of data from three independent experiments. *, P < 0.05, compared with the controls.
JunB is not involved in the HCV-induced enhancement of p16 mRNA levels
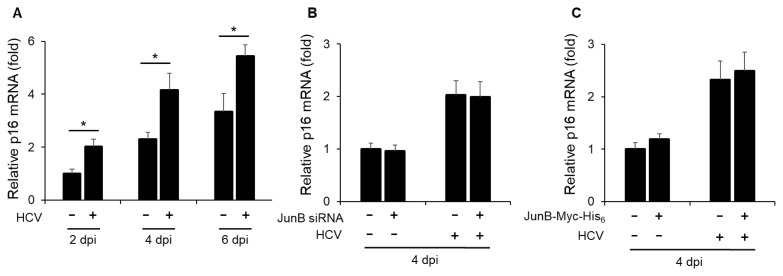
To clarify the role of JunB in HCV-associated pathogenesis, we first examined the effect of HCV infection on the mRNA levels of cyclin-dependent kinase inhibitor p16 (INK4a). Real-time RT-PCR analysis revealed a significant increase in p16 mRNA levels in HCV-infected Huh-7.5 cells at 2, 4, and 6 dpi compared to mock-infected cells (Fig. 4A). Next, to determine the effect of JunB on p16 mRNA levels, Huh-7.5 cells were infected with HCV J6/JFH1 in the presence or absence of JunB siRNA. The results showed that knockdown of JunB had no effect on the HCV-induced enhancement of p16 mRNA levels (Fig. 4B). Similarly, the overexpression of JunB also had no effect on the HCV-induced enhancement of p16 mRNA levels (Fig. 4C). These results suggest that JunB is not involved in the HCV-induced enhancement of p16 mRNA levels.
Fig. 4. JunB is not involved in the HCV-induced enhancement of p16 mRNA levels.
(A) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 2, 4, and 6 dpi, the cells were harvested, the total RNA was extracted, and mRNA levels of p16 were quantitated by real-time RT-PCR. Amounts of intracellular p16 mRNA were normalized to amounts of GAPDH mRNA. Data represent means ± SEM of data from three independent experiments. *, P < 0.05 compared with the controls. (B) Huh-7.5 cells were transfected with 40 nM JunB siRNA or control siRNA. At 24 h post-transfection, cells were infected with HCV J6/JFH1 at an MOI of 1. At 4 dpi, the cells were harvested. The total RNA was extracted and mRNA levels of p16 were quantitated by real-time RT-PCR. (C) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with either pEF1A-JunB-Myc-His6 or pEF1A-Myc-His6. At 4 dpi, the cells were harvested. The total RNA was extracted and mRNA levels of p16 were quantitated by real-time RT-PCR.
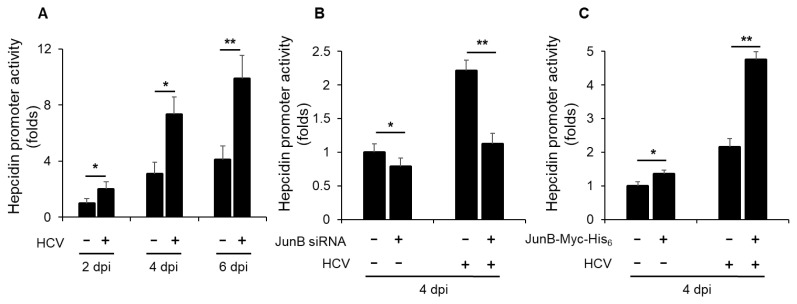
JunB is involved in HCV-induced enhancement of hepcidin promoter activity
To elucidate a role of JunB in HCV-mediated iron metabolism disorder, we first investigated the impact of HCV infection on hepcidin promoter activity. Luciferase promoter assays demonstrated a significant enhancement of hepcidin promoter activity in HCV-infected Huh-7.5 cells at 2, 4, and 6 dpi compared to mock-infected control cells (Fig. 5A). Notably, knockdown of JunB significantly disrupted the HCV-induced enhancement of hepcidin promoter activity at 4 dpi (Fig. 5B). Conversely, overexpression of JunB markedly augmented the HCV-induced enhancement of hepcidin promoter activity at 4 dpi compared to control cells (Fig. 5C). These results suggest that JunB is involved in HCV-induced enhancement of hepcidin promoter activity.
Fig. 5. JunB is involved in HCV-induced enhancement of hepcidin promoter activity.
(A) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with pGL4.10-hepcidin promoter. At 2, 4, and 6 dpi, the cells were harvested, the hepcidin promoter activities were measured by using a luciferase reporter assay. Luciferase activity was normalized to Renilla activity for each well. (B) Huh-7.5 cells were transfected with 40 nM JunB siRNA or control siRNA. At 24 h post-transfection, cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with pGL4.10-hepcidin promoter. At 4 dpi, the cells were harvested. The luciferase activity was measured and normalized to the Renilla activity. (C) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with pGL4.10-hepcidin promoter together with either pEF1A-JunB-Myc-His6 or pEF1A-Myc-His6. At 4 dpi, the cells were harvested. The luciferase activity was measured and normalized to the Renilla activity. Data represent means ± SEM of data from three independent experiments. *, P < 0.05; **, P < 0.01 compared with the controls.
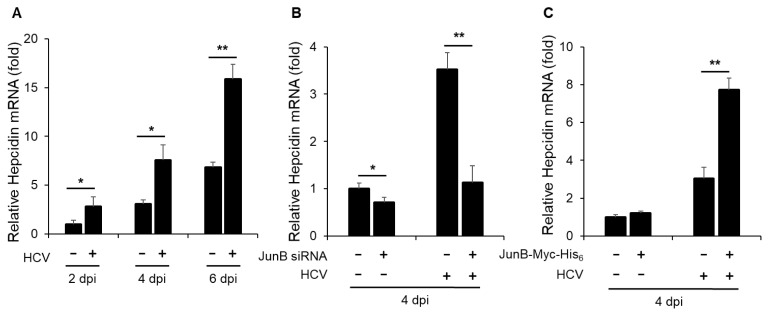
JunB is involved in HCV-induced enhancement of hepcidin mRNA levels
Next, to determine the effect of HCV infection on hepcidin mRNA levels, we performed real-time RT-PCR assays. The results revealed a significant increase in hepcidin mRNA levels in HCV-infected Huh-7.5 cells at 2, 4, and 6 dpi compared to mock-infected control cells (Fig. 6A). To further elucidate the impact of JunB on hepcidin mRNA levels, Huh-7.5 cells were infected with HCV J6/JFH1 in the presence or absence of JunB siRNA. The results demonstrated that knockdown of JunB significantly reduced the HCV-induced enhancement of hepcidin mRNA levels at 4 dpi (Fig. 6B). Conversely, overexpression of JunB further increased the HCV-induced enhancement of hepcidin mRNA levels at 4 dpi (Fig. 6C). These results suggest that JunB is involved in HCV-mediated iron metabolism disorder by activating the hepcidin promoter, thereby increasing hepcidin mRNA levels.
Fig. 6. JunB is involved in HCV-induced hepcidin mRNA levels.
(A) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 2, 4, and 6 dpi, the cells were harvested. The total RNA was extracted and mRNA levels of hepcidin were quantitated by real-time RT-PCR. (B) Huh-7.5 cells were transfected with 40 nM JunB siRNA or control siRNA. At 24 h post-transfection, cells were infected with HCV J6/JFH1 at an MOI of 1. At 4 dpi, the cells were harvested. The total RNA was extracted and mRNA levels of hepcidin were quantitated by real-time RT-PCR. (C) Huh-7.5 cells were infected with HCV J6/JFH1 at an MOI of 1. At 3 h postinfection, cells were transfected with either pEF1A-JunB-Myc-His6 or pEF1A-Myc-His6. At 4 dpi, the cells were harvested. The total RNA was extracted and mRNA levels of hepcidin were quantitated by real-time RT-PCR. Data represent means ± SEM of data from three independent experiments. *, P < 0.05; **, P < 0.01 compared with the controls.
Taken together, these results suggest that the ROS/JNK/JunB signaling pathway negatively regulates HCV replication and also contributes to iron metabolism disorder via the activation of hepcidin transcription.
DISCUSSION
In this study, we obtained results suggesting that HCV infection promotes phosphorylation of the transcription factor JunB via the ROS/JNK signaling pathway (Fig. 1). Importantly, the present study suggests that JunB plays a role in inhibiting HCV replication in both HCV genotypes 2a and 1b (Figs. 2 and 3). To explore the role of JunB in HCV-mediated iron metabolism disorder, we investigated the effect of JunB on hepcidin transcription in HCV-infected cells. The luciferase promoter assay and real-time RT-PCR analyses revealed that HCV infection significantly enhanced hepcidin promoter activity (Fig. 5A) and hepcidin mRNA levels (Fig. 6A). Furthermore, our results suggest that JunB participates in HCV-induced hepcidin promoter activity and hepcidin mRNA levels (Figs. 5B, 5C, 6B, and 6C), indicating that JunB is involved in facilitating hepcidin transcription upon HCV infection. Collectively, our results suggest that the ROS/JNK/JunB signaling pathway plays roles in inhibiting HCV replication and contributing to HCV-mediated iron metabolism disorder.
It was reported that JNK facilitates IL-1 beta-induced hepcidin transcription via the cAMP response element site B on the hepcidin promoter in hepatocytes without HCV infection (21). Our current findings demonstrated the involvement of JunB in HCV-mediated iron metabolism disorder by enhancing hepcidin transcription. To the best of our knowledge, this is the first study to elucidate the role of JunB in the HCV-mediated iron metabolism disorder.
Clinical studies have suggested a close link between HCV and iron metabolism disorder, with iron accumulation in the liver tissue of patients with chronic hepatitis C (16, 17). It is widely recognized that HCV disrupts iron metabolism by regulating hepcidin levels. Hepcidin regulates iron metabolism by interacting with ferroportin, the sole iron exporter, and promoting degradation of ferroportin, thereby controlling the primary influx of iron into plasma (27, 28). Clinical studies have reported higher levels of hepcidin in the serum of both acute and chronic HCV patients (29, 30), which is consistent with our present data demonstrating that HCV infection significantly increases hepcidin promoter activity and hepcidin mRNA levels.
Our results suggest that JunB inhibits HCV replication, although the underlying mechanism remains to be determined. A previous study has demonstrated that hepcidin inhibits HCV replication by activating the STAT3 pathway (31). Furthermore, increased hepcidin expression can lead to elevated hepatic iron storage (19, 20), and excess iron can inhibit the enzymatic activity of the HCV NS5B RNA polymerase, thereby blocking HCV replication (32, 33). Thus, we speculate that the JunB-induced enhancement of hepcidin levels plays a negative role in HCV replication by increasing the amount of iron in HCV-infected cells. We speculate that JunB may function as a cellular defense mechanism against HCV infection through the activation of JunB target genes, such as hepcidin. Further investigation is needed to explore the relationship between elevated hepcidin mRNA levels and HCV replication.
In this study, we demonstrate that the HCV-induced ROS/JNK signaling pathway activates the transcription factor JunB, which plays roles in inhibiting HCV replication and contributing to HCV-mediated enhancement of hepcidin transcription. Thus, we propose that the ROS/JNK/JunB signaling pathway negatively regulates HCV replication and also contributes to HCV-mediated iron metabolism disorder.
ACKNOWLEDGEMENTS
We are grateful to T. Suzuki (Department of Virology and Parasitology, Hamamatsu University School of Medicine, Hamamatsu, Japan) for providing pGL4.10-hepcidin promoter. We also thank Y. Kozaki for the secretarial work. This work was supported by the Program Basic and Clinical Research on Hepatitis from the Japan Agency for Medical Research and Development (AMED) under Grant no. JP 22fk0210090s1202. A.A. is supported by the Program for Nurture of Next Generation Leaders Guiding Medical Innovation in Asia of the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan.
Footnotes
AUTHOR CONTRIBUTIONS
A.A., L.D., and I.S. conceived and designed the experiments. A.A., S.Y., and Y.L. carried out most of the experiments. T.A. and C.M. assisted the constructions and the data analysis. A.A., L.D., and I.S. wrote the manuscript.
CONFLICTS OF INTEREST
The authors have no conflicts of interest to declare.
REFERENCES
- 1.World Health Organization [Internet] Hepatitis C. [cited 2023 April 22]. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c.
- 2.Llovet JM, Zucman-Rossi J, Pikarsky E, Sangro B, Schwartz M, Sherman M, et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 3.Vermehren J, Park JS, Jacobson IM, Zeuzem S. Challenges and perspectives of direct antivirals for the treatment of hepatitis C virus infection. J Hepatol. 2018;69(5):1178–1187. doi: 10.1016/j.jhep.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Heffernan A, Cooke GS, Nayagam S, Thursz M, Hallett TB. Scaling up prevention and treatment towards the elimination of hepatitis C: a global mathematical model. Lancet. 2019;393(10178):1319–1329. doi: 10.1016/S0140-6736(18)32277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19(7):837–849. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng L, Shoji I, Ogawa W, Kaneda S, Soga T, Jiang DP, et al. Hepatitis C virus infection promotes hepatic gluconeogenesis through an NS5A-mediated, FoxO1-dependent pathway. J Virol. 2011;85(17):8556–8568. doi: 10.1128/JVI.00146-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, et al. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol. 2008;82(21):10375–10385. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng L, Liang Y, Ariffianto A, Matsui C, Abe T, Muramatsu M, et al. Hepatitis C Virus-Induced ROS/JNK Signaling Pathway Activates the E3 Ubiquitin Ligase Itch to Promote the Release of HCV Particles via Polyubiquitylation of VPS4A. J Virol. 2022;96(6):e0181121. doi: 10.1128/jvi.01811-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55(3):395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 10.Zeke A, Misheva M, Remenyi A, Bogoyevitch MA. JNK Signaling: Regulation and Functions Based on Complex Protein-Protein Partnerships. Mol Biol Rev. 2016;80(3):793–835. doi: 10.1128/MMBR.00043-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B, Tournier C, Davis RJ, Flavell RA. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO J. 1999;18(2):420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001;20(19):2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 13.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 14.Passegue E, Wagner EF. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 2000;19(12):2969–2979. doi: 10.1093/emboj/19.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergman MR, Cheng S, Honbo N, Piacentini L, Karliner JS, Lovett DH. A functional activating protein 1 (AP-1) site regulates matrix metalloproteinase 2 (MMP-2) transcription by cardiac cells through interactions with JunB-Fra1 and JunB-FosB heterodimers. Biochem J. 2003;369(Pt 3):485–496. doi: 10.1042/BJ20020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hezode C, Cazeneuve C, Coue O, Roudot-Thoraval F, Lonjon I, Bastie A, et al. Liver iron accumulation in patients with chronic active hepatitis C: prevalence and role of hemochromatosis gene mutations and relationship with hepatic histological lesions. J Hepatol. 1999;31(6):979–984. doi: 10.1016/s0168-8278(99)80308-0. [DOI] [PubMed] [Google Scholar]
- 17.Silva IS, Perez RM, Oliveira PV, Cantagalo MI, Dantas E, Sisti C, et al. Iron overload in patients with chronic hepatitis C virus infection: clinical and histological study. J Gastroenterol Hepatol. 2005;20(2):243–248. doi: 10.1111/j.1440-1746.2004.03549.x. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka H, Fujita N, Sugimoto R, Urawa N, Horiike S, Kobayashi Y, et al. Hepatic oxidative DNA damage is associated with increased risk for hepatocellular carcinoma in chronic hepatitis C. Br J Cancer. 2008;98(3):580–586. doi: 10.1038/sj.bjc.6604204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt PJ. Regulation of Iron Metabolism by Hepcidin under Conditions of Inflammation. J Biol Chem. 2015;290(31):18975–18983. doi: 10.1074/jbc.R115.650150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306(5704):2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 21.Kanamori Y, Murakami M, Matsui T, Funaba M. JNK facilitates IL-1beta-induced hepcidin transcription via JunB activation. Cytokine. 2018;111:295–302. doi: 10.1016/j.cyto.2018.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76(24):13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 24.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11(7):791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami K, Ishii K, Ishihara Y, Yoshizaki S, Tanaka K, Gotoh Y, et al. Production of infectious hepatitis C virus particles in three-dimensional cultures of the cell line carrying the genome-length dicistronic viral RNA of genotype 1b. Virology. 2006;351(2):381–392. doi: 10.1016/j.virol.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 26.Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM, et al. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010;6(9):e1001087. doi: 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meynard D, Babitt JL, Lin HY. The liver: conductor of systemic iron balance. Blood. 2014;123(2):168–176. doi: 10.1182/blood-2013-06-427757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muckenthaler MU, Rivella S, Hentze MW, Galy B. A Red Carpet for Iron Metabolism. Cell. 2017;168(3):344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foka P, Dimitriadis A, Karamichali E, Kyratzopoulou E, Giannimaras D, Koskinas J, et al. Alterations in the iron homeostasis network: A driving force for macrophage-mediated hepatitis C virus persistency. Virulence. 2016;7(6):679–690. doi: 10.1080/21505594.2016.1175700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugimoto R, Fujita N, Tomosugi N, Hara N, Miyachi H, Tanaka H, et al. Impaired regulation of serum hepcidin during phlebotomy in patients with chronic hepatitis C. Hepatol Res. 2009;39(6):619–624. doi: 10.1111/j.1872-034X.2009.00497.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu H, Trinh TL, Dong H, Keith R, Nelson D, Liu C. Iron regulator hepcidin exhibits antiviral activity against hepatitis C virus. PLoS One. 2012;7(10):e46631. doi: 10.1371/journal.pone.0046631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fillebeen C, Rivas-Estilla AM, Bisaillon M, Ponka P, Muckenthaler M, Hentze MW, et al. Iron inactivates the RNA polymerase NS5B and suppresses subgenomic replication of hepatitis C Virus. J Biol Chem. 2005;280(10):9049–9057. doi: 10.1074/jbc.M412687200. [DOI] [PubMed] [Google Scholar]
- 33.Fillebeen C, Pantopoulos K. Iron inhibits replication of infectious hepatitis C virus in permissive Huh7.5.1 cells. J Hepatol. 2010;53(6):995–999. doi: 10.1016/j.jhep.2010.04.044. [DOI] [PubMed] [Google Scholar]