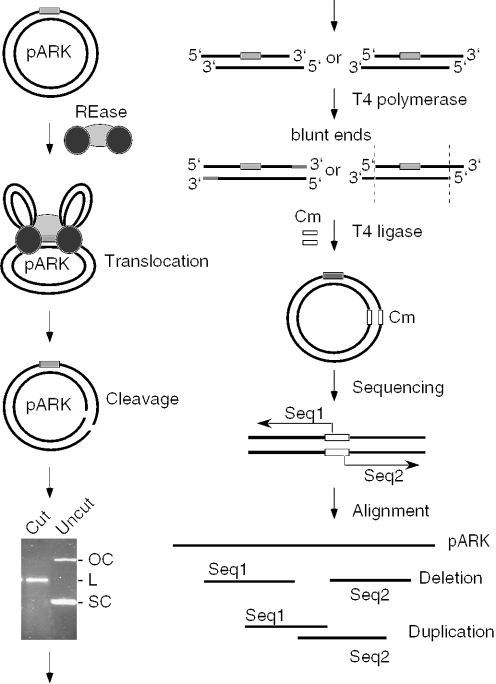
Figure 1.
Experimental strategy for the analysis of the DNA ends produced by Type I restriction enzymes. Circular plasmid DNA (black lines) containing a single recognition site (gray box) is cleaved by a Type I restriction enzyme (REase) that is represented as one light gray oval (methylase) and two dark gray ovals (HsdR subunits). The linearized DNA is purified from an agarose gel and treated with T4 DNA polymerase to remove possible 3′-overhangs or to fill in 5′-overhangs. The resulting blunt-ended DNA fragments are ligated with a fragment carrying a chloramphenicol (Cm) resistance marker (open box) and transformed into bacteria. Plasmid DNA from individual clones is isolated and the sequence of the ends of cloned Type I restriction products is determined using primers originating in the Cm gene. The obtained sequences are aligned with the original pARK sequence. Deletions at the ends of cloned DNA fragments suggest the presence of 3′-overhangs, while sequence duplications indicate 5′-overhangs. No change in the sequence with respect to pARK indicates the presence of blunt ends.