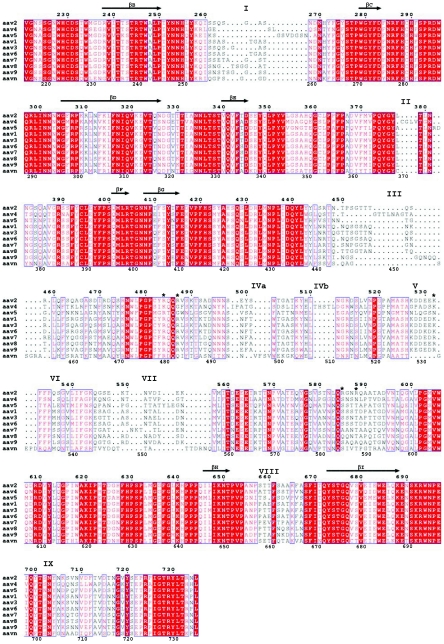
FIG.3.
Structural alignment of AAV1-9 and AAAV (AAVN). The alignment was generated based on superimposition of the atomic coordinates of AAV2 (68), the pseudoatomic structures of AAV4 and AAV5 (63) built into their reconstructed cryo-EM densities, and structural models of AAV1, AAV3, AAV6-9, and AAAV generated with the Swiss Model program (50), with AAV2 as a template. The format was output by use of the program ESPript (26). Identical residues are white characters in red boxes; homologous residues are red characters in white boxes; differing residues are black characters. The numbering at the top of the alignment is based on AAV2 VP1 sequence numbers, while that at the bottom is based on AAV4 VP1 sequence numbers. Variable surface loop regions are labeled I to IX. Residues R484, R487, K532, R585, and R588 (AAV2 VP1 numbering), implicated in AAV2 heparin binding (38, 46), are indicated by asterisks. The residues within the eight core β-strands that form the contiguous parvovirus capsid are indicated by arrows above the residues.