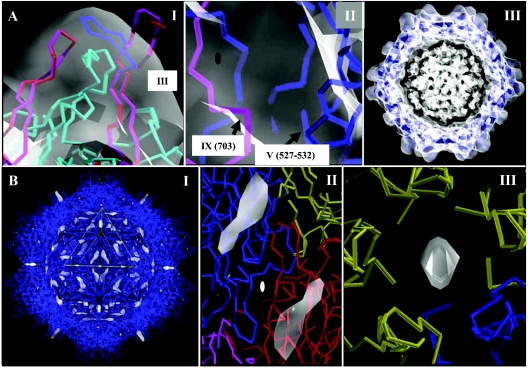
FIG. 4.
Pseudoatomic model building for the AAV4 capsid. (A) Panel 1 shows the C-α traces of the AAV4 monomer (in magenta) generated by Swiss Model (50) that required adjustment (in blue) in the program O (34) to fit inside the reconstructed density (in transparent white contoured at a σ of 1.0) of the mounds surrounding the icosahedral threefold axes and the starting AAV2 crystal structure model (in red) (68). An intervening loop from a threefold related VP3 subunit is shown in cyan. Variable region III is labeled. Panel II shows C-α traces for loop regions in a reference AAV4 VP3 (in blue) and a twofold related monomer (in magenta), residues 527 to 532 and near 703, respectively, that could not be adjusted to fit into the cryo-EM density envelope (in transparent white contoured at a σ of 1.0) at 13-Å resolution. These regions are labeled V and IX, respectively, as in Fig. 3 to 6. The approximate icosahedral twofold axis is indicated by the filled oval. Panel III shows the C-α traces for 60 AAV4 VP3 monomers (in blue) fitted into the cryo-EM density (in transparent white contoured at a σ of 1.0). The 60 VP3 molecules were generated by matrix multiplication of the coordinates of a reference VP3, in a standard orientation, with 60 icosahedral symmetry operators after model refinement in a crystallography and NMR system (CNS) program (13). The map is sectioned at the center to show the protein envelope. The density in the middle of the particle, at the same contour level as the protein envelope, could not be modeled as a protein or nucleic acid. The view is approximately down an icosahedral twofold axis. (B) Difference map density subtracting the VP3 model from the cryo-EM density. Panel I shows the 60 AAV4 VP3 monomers (in blue) and the positions of difference density (in transparent white) at a contour level of 2.2 σ. A viral asymmetric unit is shown in the yellow triangle, bounded by two threefold axes (bottom right and left edges) and a fivefold axis (top edge), with a twofold axis in the middle. The view, down an icosahedral twofold axis, is rotated 90° compared to the twofold views shown in Fig. 2. Panels II and III show close-up views of the difference densities (in transparent white) viewed down an icosahedral twofold axis (from the interior of the capsid) and in the channel at the icosahedral fivefold axes (viewed from the capsid exterior). The reference monomer is in blue in both panels, the twofold monomer is in red in panel II, and the fivefold monomers are in yellow in panel III.