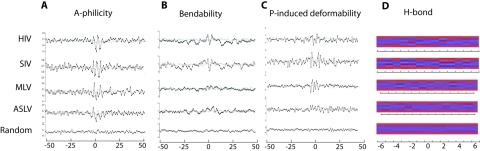
FIG. 2.
DNA physical properties of retroviral integration sites. (A) A-philicity, (B) DNA bendability, and (C) protein-induced deformability scores were calculated as described for the 100-bp sequence around the integration site, with the center of the duplicated integration target site marked as position 0 bp. Nucleotide positions are numbered by − for the 5′ end of the integration site and + for the 3′ end of the integration site. A-philicity scores and protein-induced deformability scores were calculated based on dinucleotide frequency. DNA bendability was calculated based on trinucleotide frequency. A set of 500 computer-generated random integration sites in human genome was used as a random control. The scores of individual integration sites at each base position were averaged for each virus (black line) or the random sites (gray line) and plotted. (D) Hydrogen bonding patterns for retroviral integration sites and random control sites were drawn with HbondView software as described. Position 0 represents the center of the target site duplication, and only patterns for the surrounding 12 bp are shown. Each base pair has six potential hydrogen bond sites in the major groove of DNA. The potentials for hydrogen bond donor or acceptor are color coded as follows: red represents a potential acceptor site, blue represents a potential donor site, and gray indicates a site that can be neither a donor nor an acceptor. The patterns shown here represent the average potential at a specific site over all aligned integration sites of a specific virus or random sites. Palindromic patterns centered on the virus-specific target site duplication are apparent.