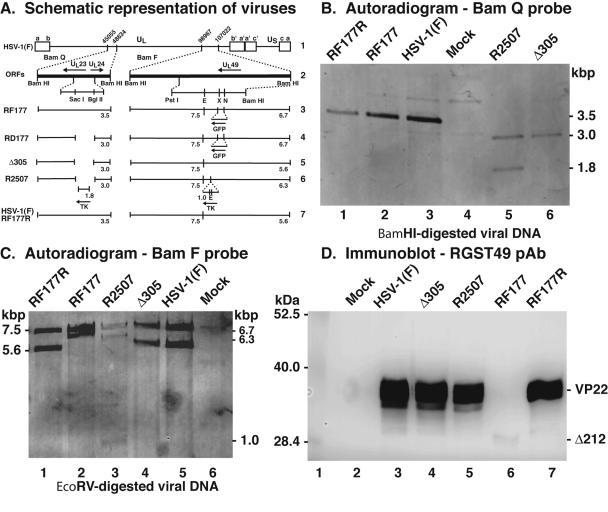
FIG. 6.
Repair of HSV-1(RF177). (A) Schematic representation of viruses used in this study. Line 1, wild-type HSV-1 in the prototype orientation. The unique long (UL), unique short (US), terminal repeat segments (a, b, c, a′, b′, and c′), and relevant nucleotide numbers are indicated. Line 2, relevant predicted open reading frames in the BamQ and BamF fragments of the viral genome. Locations of SacI, BglII, PstI, EcoRV (E), XhoI (X), and NsiI (N) restriction sites are indicated. Lines 3 to 7, expected fragments (numbers are kilobase pairs) following hybridization with either BamQ or BamF probes for RF177, RD177, Δ305, R2507, HSV-1(F), and RF177R viruses. (B and C) Autoradiographic images of viral DNAs. Southern hybridizations of BamHI-digested viral DNA were performed using BamQ- or BamF-specific probes as described in Materials and Methods. The sizes of viral DNA fragments hybridizing to the probes are indicated in the margins. (D) Immune reactivities of infected cell proteins. Whole-cell extracts of mock-, HSV-1(F), Δ305, R2507, RF177, HSV-1(F), and RF177R-infected Vero cells were prepared at 24 hpi, separated in a denaturing gel, transferred to nitrocellulose, and probed with anti-VP22 polyclonal antibody (RGST49 pAb). The locations of molecular mass markers (in kilodaltons) are indicated in the left margin. Δ212 is the truncated form of VP22 synthesized by RF177.