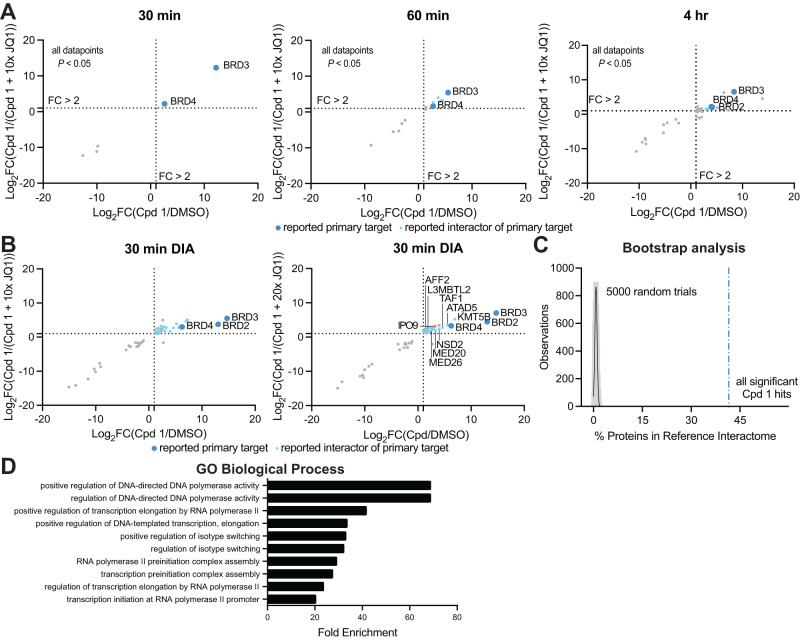
Fig. 2. The BioTAC system enables rapid, accurate small molecule interactome-ID.
A Time-course proteomics using data-dependent acquisition methods of streptavidin-enriched biotinylated proteins isolated from HEK293 cells transiently transfected with miniTurboFKBP12F36V and treated with the indicated compounds and 100 µM biotin, demonstrating enrichment and competition of known direct targets (30 min) and complexed proteins (60 min, 4 h). Only proteins with P-value < 0.05 (two-sample moderated T-test) in both conditions depicted. FC = fold-change, complete datasets in Supplementary Data 1, plotted individually in Supplementary Fig. 4. High-confidence hits are defined as those that are enriched >2-fold in both Cpd 1/ DMSO and Cpd 1/Cpd 1 + 10x (+)-JQ1, where P < 0.05, plotted upper-right quadrant. B Proteomics using data-independent acquisition methods of streptavidin-enriched biotinylated proteins isolated from HEK293 cells transiently transfected with miniTurboFKBP12F36V and treated with the indicated compounds and 100 µM biotin for 30 min, demonstrating enrichment and competition of known direct targets and complexed proteins. Only proteins with P-value < 0.05 (two-sample moderated T test) in both conditions depicted. FC = fold-change, complete datasets in Supplementary Data 1. C Percent enrichment of reference interactors (Lambert et al.21) in the hits identified from datasets depicted in panels A and B vs. the percent enrichment of reference interactors in 5000 random protein sets of equivalent size from the human transcriptome, showing significant enrichment by Cpd 1 (>41 standard deviations away from mean of random chance). D Gene Ontology analysis, showing significant enrichment of biological processes associated with known BET-protein function in the datasets depicted in panels A and B. Source data are provided as a Source Data file.