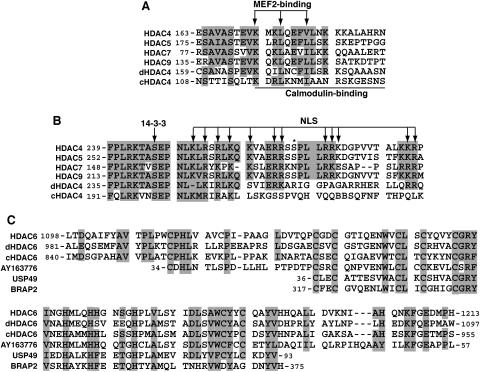
FIG. 2.
(A and B) Sequence alignment of the MEF2-binding motif (A) and the NLS (B) of class IIa HDACs. Residues invariant and highly conserved among at least four sequences are shaded. Arrows, residues known to be essential for the NLS function, 14-3-3 association, or MEF2 binding; solid line, region critical for Ca2+/calmodulin binding; asterisk, potential Dyrk1B phosphorylation site. (C) Sequence similarity between the HUB finger of HDAC6 and motifs in other proteins. Identical and highly conserved residues are shaded. Only the central region of the HUB finger is similar to USP49 and BRAP2. Residues invariant and highly conserved among at least 75% of the sequences are shaded.