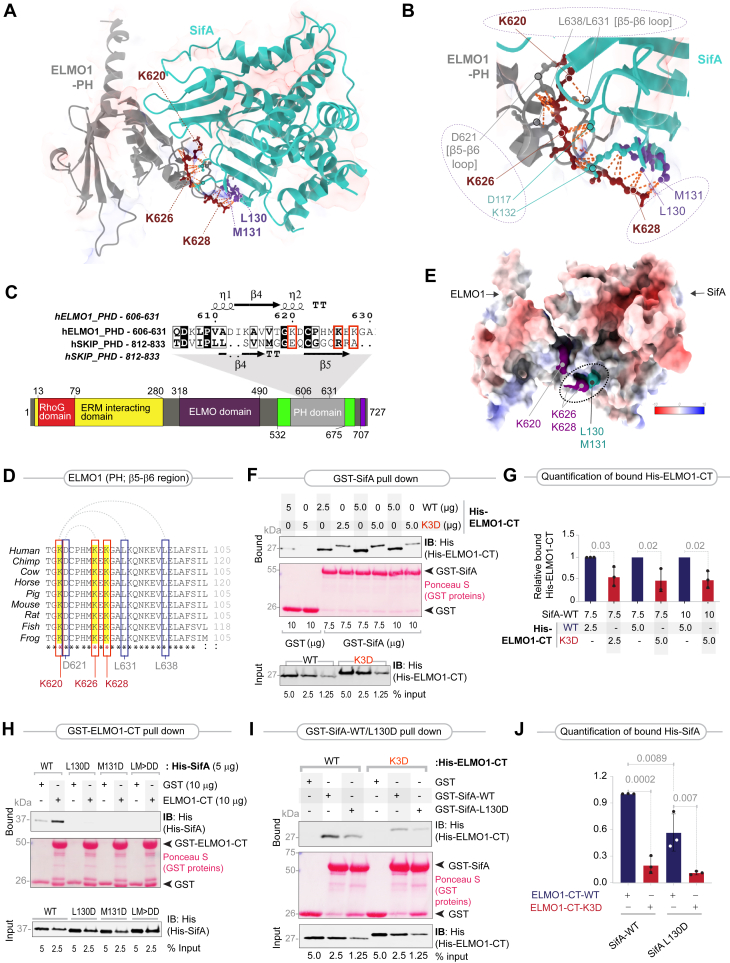
Figure 2.
Characterization of the ELMO1–SifA interface.A, homology model of the ELMO1 (gray)–SifA (turquoise) complex, generated by superimposing the solved structure of the ELMO1-PH (PDB code: 2VSZ) on that of the SKIP-PH in complex with SifA (PDB code: 3CXB). The executive RMSD of the model is 1.528 (see Fig. S1 for the workflow used to generate the homology models and select the fittest model with the most optimal parameters). The model is annotated with key amino acids that are predicted to form the interface. On ELMO1, these amino acids were identified as an evolutionarily conserved polar hot spot of three lysines within the β5–β6 loop, and their key intramolecular and intermolecular contacts on SifA are annotated. See Supporting Information Data 1 for a complete catalog of the intermolecular and intramolecular contacts of the highlighted residues. The distance between the residues calculated for contacts was within 4.0 A. See Fig. S2 for the position of the WxxxE motif relative to the ELMO1–SifA interface. B, a magnified view of the key residues participating at the ELMO1 (gray)–SifA (turquoise) interface. Three major clusters of intermolecular and intramolecular interactions of the lysine triad are annotated with interrupted circles/ovals. Lys(K)628 on (ELMO1) primarily engages via strong polar contacts with Met(M)131 and Leu(L)130 on SifA. K626 on (ELMO1) makes an intramolecular contact with D621, a residue within the β5–β6 loop; it is also juxtaposed with K132 and forms a “charge-neutralizing” salt bridge with Asp(D)117 on SifA. K620 on (ELMO1) appears to primarily bind L631 and L638, which are key residues within the β5–β6 loop. C, top, an alignment of the sequences of the PH domains of ELMO1 and SKIP is shown, along with secondary structures. Conserved residues are shaded in black; similar residues are boxed. Three lysine residues on ELMO1 that correspond to the structurally resolved contact sites of SKIP for SifA are marked with red boxes. See also Fig. S3A for an extended alignment. Bottom, a domain map of ELMO1. D, an alignment of the β5–β6 loop of ELMO1 showing that the lysine triad highlighted with red box in (C) is conserved across diverse species. Intraloop interactions are indicated with interrupted arcs on top. Leu(L) and Asp(D) residues that are engaged in these interactions are also conserved and highlighted with blue boxes. See also Fig. S4 for extended alignment. E, the panel displays APBS (Adaptive Poisson–Boltzmann Solver)-derived surface electrostatics for an all-side chain model of the ELMO1–SifA cocomplex, as visualized using Chimera. Volume surface coloring was set in the default range of −10 (red), through 0 (white), to +10 (blue) kT/e, where negatively charged surfaces are red (−10 kT/e) and positively charged surfaces are blue (+10 kT/e). In the most energetically favorable orientation, charged residues Lys(K)628 and Lys(K)626 on (ELMO1) bring hydrophobic residues Met(M)131 and Leu(L)130 on SifA into proximity (marked by an oval). See Fig. S5 for additional views. F, recombinant WT or K3D mutant His-ELMO1-CT proteins (input) were used in pulldown assays with immobilized GST alone or GST-SifA (∼7.5 or 10 μg). Bound ELMO1 was visualized by immunoblotting using an anti-His (ELMO1) antibody. GST proteins are visualized by Ponceau S staining. The K3D mutant displayed slower electrophoretic mobility compared with the WT ELMO1 protein consistently in both reducing and nonreducing gels, and regardless of whether it was expressed in bacteria as recombinant proteins or expressed in mammalian cells, suggesting it is likely to be due to the introduction of negative charge in the form of three aspartates. G, quantification of immunoblots in (F). Results are displayed as mean ± SD (n = 3 independent replicates). Statistical significance was determined using an unpaired t test. H, equal aliquots (input) of recombinant His-SifA and its mutants (L130D, M131D, and LM-DD) were used in pulldown assays with immobilized GST alone or GST-ELMO1-CT (10 μg). Bound SifA proteins were visualized by immunoblotting using an anti-His antibody. GST proteins are visualized by Ponceau S staining. I, recombinant WT or K3D mutant His-ELMO1-CT proteins were used in pulldown assays with GST or GST-SifA (WT or L130D mutant). Bound ELMO1 was visualized by immunoblotting using an anti-His antibody. GST proteins are visualized by Ponceau S staining. J, quantification of immunoblots in (I). Results are displayed as mean ± SD (n = 3 independent replicates). Statistical significance was determined using an unpaired t test. ELMO1, Engulfment and Cell Motility protein 1; GST, glutathione-S-transferase; PDB, Protein Data Bank.