Abstract
Background and Objectives
The spinocerebellar ataxias (SCAs) are a genetically heterogeneous group of neurodegenerative disorders generally caused by single nucleotide variants (SNVs) or indels in coding regions or by repeat expansions in coding and noncoding regions of SCA genes. Copy number variants (CNVs) have now also been reported for 3 genes—ITPR1, FGF14, and SPTBN2—but not all SCA genes have been screened for CNVs as the underlying cause of the disease in patients. In this study, we aim to assess the prevalence of CNVs encompassing 36 known SCA genes.
Methods
A cohort of patients with cerebellar ataxia who were referred to the University Medical Center Groningen for SCA genetic diagnostics was selected for this study. Genome-wide single nucleotide polymorphism (SNP) genotyping was performed using the Infinium Global Screening Array. Following data processing, genotyping data were uploaded into NxClinical software to perform CNV analysis per patient and to visualize identified CNVs in 36 genes with allocated SCA symbols. The clinical relevance of detected CNVs was determined using evidence from studies based on PubMed literature searches for similar CNVs and phenotypic features.
Results
Of the 338 patients with cerebellar ataxia, we identified putative clinically relevant CNV deletions in 3 patients: an identical deletion encompassing ITPR1 in 2 patients, who turned out to be related, and a deletion involving PPP2R2B in another patient. Although the CNV deletion in ITPR1 was clearly the underlying cause of SCA15 in the 2 related patients, the clinical significance of the deletion in PPP2R2B remained unknown.
Discussion
We showed that CNVs detectable with the limited resolution of SNP array are a very rare cause of SCA. Nevertheless, we suggest adding CNV analysis alongside SNV analysis to SCA gene diagnostics using next-generation sequencing approaches, at least for ITPR1, to improve the genetic diagnostics for patients.
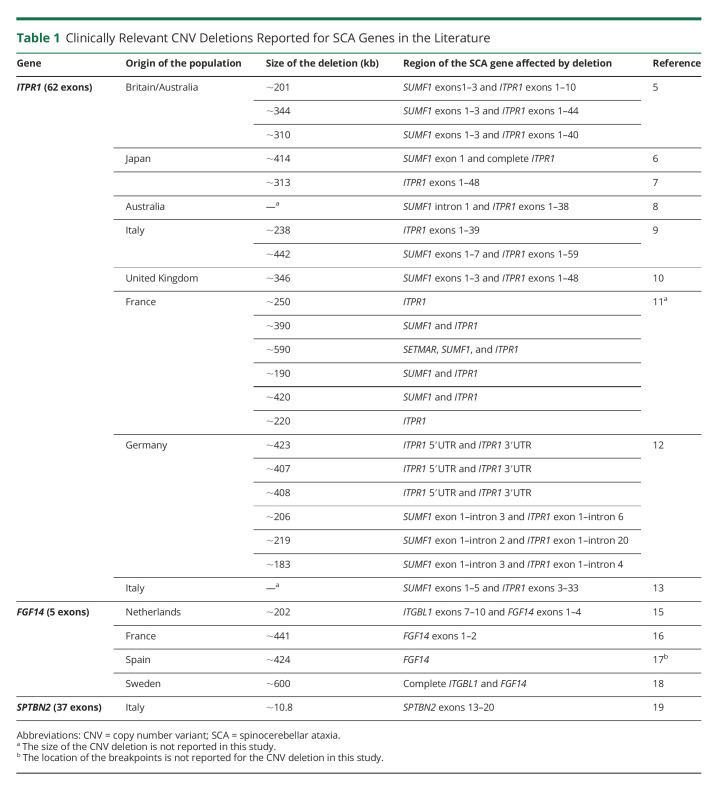
Autosomal dominant cerebellar ataxias, known as spinocerebellar ataxias (SCAs), are a clinically and genetically heterogeneous group of neurodegenerative disorders characterized by loss of balance, gait alteration, dysarthria, and incoordination.1 To date, about 50 SCA types have been identified based on the disease gene or the chromosomal location if the causal gene has not yet been identified.2 SCA types can further be distinguished by the type of genetic aberration. SCA is mainly caused by CAG repeat expansions in the coding parts of 6 genes (SCA1–3, 6, 7, and 17), leading to an elongated polyglutamine stretch in their corresponding proteins.3 Two other causes of SCA are repeat expansions in noncoding regions (SCA8, 10, 12, 31, 36, and 37) and single nucleotide variants (SNVs) or indels within coding regions (SCA5, 11, 13–15, 19, 21, 23, 26–28, 34, 35, 38, and 40–49) (eTable 1, links.lww.com/NXG/A573).2,3 Clinically relevant copy number variants (CNVs) (>1 kb)4 have been reported for only 3 SCA genes.5-19 CNV deletions encompassing ITPR1, which codes for inositol-triphosphate receptor type 1, were first identified in an Australian family with SCA15, and more studies subsequently reported CNV deletions in the ITPR1 locus in patients with different ethnic backgrounds.5-14 Second, several studies have also reported CNV deletions encompassing the FGF14 locus, which encodes fibroblast growth factor 14, in SCA27 cases.15-18 Finally, an intragenic CNV deletion was recently found in SPTBN2, which codes for b-III spectrin protein, involved in SCA5.19 Detailed descriptions of these CNV deletions can be found in Table 1.
Table 1.
Clinically Relevant CNV Deletions Reported for SCA Genes in the Literature
Unfortunately, there is still a relatively large number of patients with SCA for whom no genetic diagnosis can be made after routine genetic testing for coding repeat expansions and SNV/indels in the known SCA genes. Noncoding repeat expansions or CNVs in known SCA genes are currently not tested in genetic diagnostics in the Netherlands and might explain the disease in some patients. Although a few studies have reported cases with CNV deletions encompassing an SCA gene, not all SCA genes have been screened for CNVs as the underlying cause of SCA. Therefore, to study whether CNVs are more often the underlying cause of SCA, we performed a genome-wide single nucleotide polymorphism (SNP) array CNV analysis on a cohort of 338 patients with cerebellar ataxia and searched for clinically relevant CNVs in 36 genes with allocated SCA symbols (eTable 1, links.lww.com/NXG/A573).2 Our analysis revealed that CNVs in SCA genes, at least those detectable with SNP array, are very rare, but smaller CNVs may still represent an underlying cause of the disease. We recommend implementing CNV analysis, for in any case ITPR1, to existing next-generation sequencing (NGS) diagnostic procedures to improve the genetic diagnostics for patients.
Methods
Patient Selection
We studied 338 Dutch patients with cerebellar ataxia who were referred to the laboratory of the Department of Genetics, University Medical Center Groningen, Groningen, the Netherlands for SCA genetic diagnostics. At the moment of inclusion in the study, all patients remained without a genetic diagnosis, and for the majority of patients, repeat expansions in the most frequent SCA genes (SCA1–3, 6, 7, and 17) were excluded from the study. For this study, we only tested for the prevalence of CNVs in the SCA genes to keep the analysis in line with the original diagnostic request, and thus, no additional informed consent was requested. Clinical information about patients carrying CNVs was retrospectively collected from the clinical reports.
SNP Array CNV Analysis
Genome-wide SNP genotyping was performed using the Infinium Global Screening Array-24 v3.0-EA-MD (Illumina, San Diego, CA), as per the manufacturer's protocol. Data were processed using our in-house pipelines (github.com/molgenis/AGCT and github.com/molgenis/GAP) to produce genotypes and calculate log R ratios and B allele frequencies (BAFs). The log R ratio for each SNP represents the normalized signal intensity, which is the log2 of the ratio of the observed signal and the expected signal for 2 genomic copies.20 The fraction of SNPs that support a nonreference allele at a given position is known as the BAF (the number of times that allele A and allele B are found at each position). Expected values for BAFs are 0 for AA, 0.5 for AB, and 1 for BB, with ∆BAF representing the estimated deviation from the expected AB value.
Processed data were uploaded into NxClinical version 5.0 software (BioDiscovery, El Segundo, CA) to perform CNV analysis and visualize identified CNVs in the 36 genes with allocated SCA symbols (eTable 1, links.lww.com/NXG/A573). To exclude artifacts due to technical errors, we selected only samples with an SD <0.25 and CNVs containing at least 10 subsequent SNPs for further analysis. For deletions and duplications, only CNVs larger than 50 kb and 200 kb were selected, respectively. CNVs were visually inspected for clearly aberrant log2 ratios and BAFs. To filter out benign variants, we compared the CNVs to an in-house genome-wide CNV database produced for 3,280 healthy Dutch controls using the same array. Only CNVs present in <1% of the control group were considered potentially clinically relevant. The clinical relevance of CNVs was determined using evidence from studies based on a PubMed literature search for similar CNVs and phenotypic features.
Standard Protocol Approvals, Registrations, and Patient Consents
The SNP array CNV analysis presented in this study is in line with the original request for diagnostic testing, and therefore, no additional informed consent was requested.
Data Availability
The data presented in this article cannot be made publicly available as it considers patient information. To protect patient privacy, access to the data can only be made by request from the corresponding author.
Results
For this study, we excluded 46 low-quality samples (>0.25 SD). In the remaining 292 samples, we identified CNV deletions in 3 patients (1%): an identical deletion encompassing ITPR1 and SUMF1 in 2 patients and a deletion involving PPP2R2B in another patient. No other CNVs were detected in the other 34 SCA genes. Below, we describe the respective CNV deletions and the corresponding clinical features of the patients. No CNV duplications were identified in the patients encompassing any of the SCA genes.
ITPR1 and SUMF1 Deletion
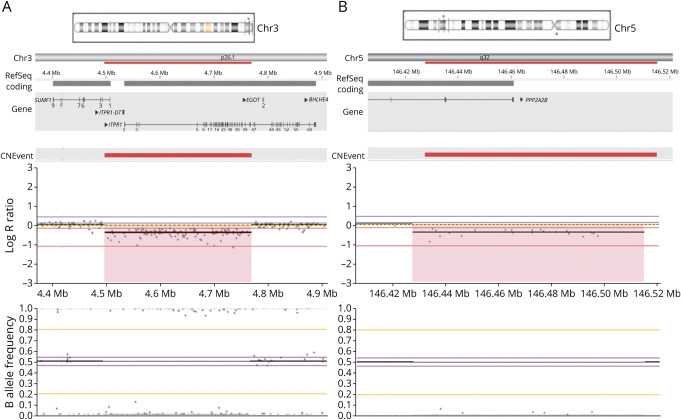
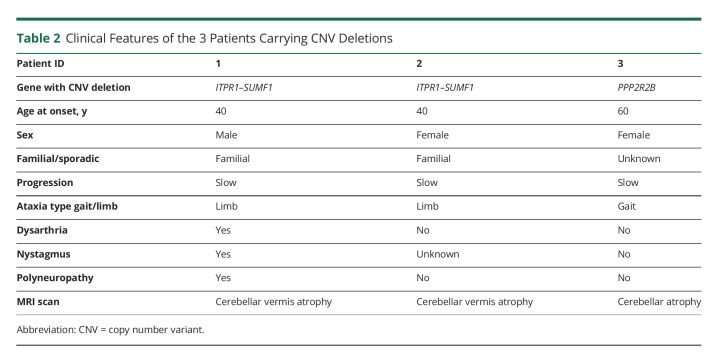
CNV analysis showed a deletion of chromosome 3p26.1 with a minimum size of 260.8 kb (108 SNP probes, GRCh37:4,503,353–4,764,171) in 2 patients. The deletion includes exons 1–41 of ITPR1 (NM_001168272.2) and exons 1 and 2 of SUMF1 (NM_182760) (Figure 1A). Examination of the SNPs flanking the deleted region allowed us to delimit the deletion to 12.3 kb on the centromeric side (between rs137852850 and rs17629635) and 10.6 kb on the telomeric side (between rs9311400 and rs61757111). Array data for this locus from the in-house control population did not show deletions affecting ITPR1 or SUMF1, whereas previous studies have reported heterozygous deletions encompassing ITPR1 as the underlying cause of SCA15 (Table 1). Given the existing literature, the CNV deletion encompassing ITPR1 and SUMF1 identified in the 2 patients can be considered pathogenetic, and subsequently, the patients could be diagnosed with SCA15. To confirm and expand on the SCA15 phenotypic spectrum, we collected available information from the clinical reports of the 2 patients (1 and 2) (Table 2). Unexpectedly, the 2 patients turned out to be a father and his daughter. Both cases had a similar age at onset (∼40 years), presented with limb ataxia, and showed signs of cerebellar vermis atrophy in their MRI scan. Dysarthria and polyneuropathy were present only in the father. Nystagmus was also present in the father but not examined for the daughter.
Figure 1. CNV Deletions in ITPR1 and PPP2R2B.
Analysis and visualization was performed using NxClinical software. (A) CNV deletion found in patients 1 and 2 (father and daughter). The deletion, located on chromosome 3p26.1, includes ITPR1 (exons 1–41) and SUMF1 (exons 1 and 2) and has a minimum size of 260.8 kb (108 SNP probes). (B) CNV deletion found in patient 3. The deletion is located on 5q32 with a minimum size of 63.6 kb (21 SNP probes), which includes the first 2 exons of PPP2R2B. The red deleted regions show homozygosity and low copy number. The upper plot shows low values for log R ratio (<0), indicating a CNV deletion. In the lower plot, the B allele frequency is not clustering around 0.5 due to the deletion of 1 copy of the corresponding gene, fitting a heterozygous pattern. Each SNP is shown as a dot. CNV = copy number variant; SNP = single nucleotide polymorphism.
Table 2.
Clinical Features of the 3 Patients Carrying CNV Deletions
PPP2R2B Deletion
We found a CNV deletion in a female patient on 5q32 with a minimum size of 63.6 kb (21 SNP probes, GRCh37:146,434,146–146,497,842), comprising the first 2 exons of PPP2R2B (NM_181677). We delimited the deletion to 13 kb on the telomeric side (between rs114419691 and rs720305) and 34.8 kb on the centromeric side (between rs76729368 and rs7733882) (Figure 1B). Deletions involving PPP2R2B were absent in the array data of the in-house control population, and only repeat expansions in the 5'UTR of PPP2R2B have been reported to cause SCA12.21 To determine whether the clinical features of the patient matched with the known features of SCA12, we collected all the available clinical information of the patient. The patient (3) presented with gait ataxia, but no signs of dysarthria, nystagmus, and polyneuropathy were present at the time of examination at age 60 years. MRI scan showed cerebellar atrophy (Table 2). During the course of our study, this patient received a genetic diagnosis of SCA3 with an ATXN3 CAG repeat expansion of 65 repeats.
Discussion
We systematically searched for clinically relevant CNVs in 36 known SCA genes in a cohort of patients with cerebellar ataxia. Using a genome-wide SNP array, we identified CNV deletions in 3 of 292 cases (1%): a ∼260.8 kb deletion involving ITPR1 and SUMF1 in 2 related cases and a ∼63.6 kb deletion in PPP2R2B in another case. Although heterozygous CNV deletions encompassing ITPR1 and SUMF1 have previously been reported to cause SCA15, CNVs in PPP2R2B have not been reported as the underlying cause of SCA12.
Heterozygous CNV deletions encompassing ITPR1 and SUMF1 were initially found in 3 families after identification of a spontaneous in-frame deletion in Itpr1 in mice with a movement disorder.5 Following this discovery, studies using different molecular techniques (e.g., DNA microarrays or multiplex ligation-dependent probe amplification) have reported variable CNV deletions encompassing ITPR1 and (in most cases) part of the neighboring gene SUMF1, as shown in Figure 2.6-13 One study also reported a large CNV deletion encompassing ITPR1 and SUMF1 and another neighboring gene, SETMAR (Figure 2).11 Of interest, heterozygous carriers of SUMF1 with a deficiency of SUMF1 do not have any movement disorder, and thus, it has been postulated that the heterozygous deletion of ITPR1, and not SUMF1, is the cause of SCA15.9 Accordingly, deletion of 1 copy of ITPR1 leads to lower mRNA levels and ultimately reduced ITPR1 protein in the cells of patients with ITPR1 deletions, and thus, the haploinsufficiency of ITPR1 is very likely the underlying disease mechanism.9,10 ITPR1 mediates Ca2+ release from the endoplasmic reticulum in different neurons, particularly Purkinje cells that are mainly affected in SCA.6 Purkinje cells are thus vulnerable to gene dosage of ITPR1, and haploinsufficiency of ITPR1 leads to aberrant intracellular Ca2+ homeostasis and dysfunction of Purkinje cells that ultimately causes their degeneration.6 Based on what has been reported before, the heterozygous ITPR1 deletion found in our study must be the underlying cause of SCA in the 2 related patients, and thus, the patients are diagnosed with SCA15. In addition, the grandfather of this family also had the disease, but a DNA sample was not available to perform a genetic test (for family pedigree, see eFigure 1A, links.lww.com/NXG/A573). A previous study on a large cohort of patients with dominant ataxia showed homogeneous clinical presentation in all patients with SCA15 carrying ITPR1 deletion.11 However, the 2 patients reported in our study were not expressing all the associated clinical features of SCA15 including polyneuropathy and dysarthria. Thus, variability in the manifestation of SCA15 disease seemingly exists.
Figure 2. Analysis of ITPR1, SUMF1, and SETMAR CNV Deletions From Previous Studies and the Present Study.
,The gene structures of SETMAR, SUMF1, and ITPR1 are shown at the top. Black lines below indicate previously reported deletions. Dotted black lines indicate that only the deleted genes and not the location of the breakpoints are reported for the CNV deletions identified in the respective study. The red line shows the deleted region in this study. CNV = copy number variant.
The clinical relevance of the 63.6-kb deletion encompassing the first 2 exons of PPP2R2B in 1 patient who also received a genetic diagnosis for SCA3 remains unknown. PPP2R2B encodes for a brain-specific regulatory subunit of protein phosphatase 2A (PP2A). PP2A is a serine/threonine phosphatase that is involved in regulation of protein phosphorylation. Only expansions of the CAG repeats located within the 5′UTR of an isoform of PPP2R2B, the presumed promoter region of the gene, are reported to cause SCA12.21 However, the functional consequence and thus the underlying disease mechanism of the noncoding CAG expansion in PPP2R2B are unknown.21
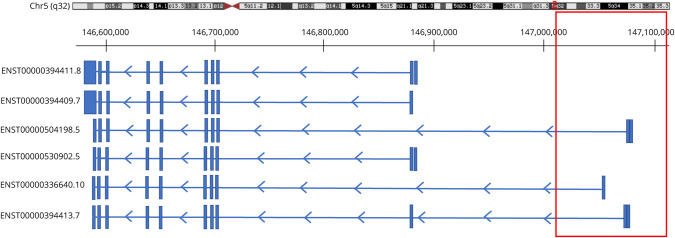
Based on the GTEx portal V8 (gtexportal.org), 6 PPP2R2B transcripts are expressed in the cerebellum: ENST00000394411.8, ENST00000504198.5, ENST00000530902.5, ENST00000336640.10, ENST00000394413.7, and ENST00000394409.7. The CNV deletion we report in PPP2R2B overlaps with exons of 3 of the transcripts: it encompasses the first exon of transcript ENST00000336640.10 and the first 2 exons of ENST00000504198.5 and ENST00000394413.7 (Figure 3). In the GTEx portal, ENST00000504198.5 has the highest expression in the cerebellum, and the deletion of its first 2 exons and the upstream 5′UTR, which likely contains important elements for gene expression, might have an effect on the expression levels of PPP2R2B and may lead to haploinsufficiency. However, the disease mechanism behind the repeat expansion in PPP2R2B is not yet known, so whether haploinsufficiency can lead to SCA12 remains to be determined.
Figure 3. Schematic Presentation of PPP2R2B Transcripts Expressed in the Cerebellum and the Location of the CNV Deletion Presented in This Study.
The red box indicates the region deleted in our patient. This CNV deletion is located at the telomeric side in an intergenic region and at the centromeric side in intron 1 of ENST00000336640.10 and intron 2 of ENST00000504198.5 and ENST00000394413.7. The CAG repeat expansion known to cause SCA12 is upstream of the first exon of ENST00000530902.5 (NR_073526).24 ENST00000504198.5 has the highest expression in the human cerebellum. CNV = copy number variant.
Based on the available clinical information, our patient (3) with the PPP2R2B deletion did not present with early-onset upper limb action tremor, which is a unique feature of SCA12 compared with other SCA types.22 With the progression of the disease, patients with SCA12 also develop additional features such as gait ataxia, head tremor, dysarthria, nystagmus, bradykinesia, hyperreflexia, and cognitive disturbances.22,23 However, in our patient, dysarthria and nystagmus were also not present. The patient seems to be a de novo case, as no other family members were found to be affected (eFigure 1B). For this patient, an established SCA3 genetic diagnosis and the absence of SCA12 clinical features support the idea that the PPP2R2B deletion we identified has a clinically insignificant effect. Moreover, the CNV deletion does not overlap with the PPP2R2B region that carries the known disease-causing CAG repeat expansion located upstream of the first exon of ENST00000530902.5 (NR_073526) (Figure 3).24 Based on these findings, the PPP2R2B deletion seems to be a rare benign variant. However, genetic modifiers—variants that modify the outcome of the main disease-causing variant—have been reported for rare mendelian diseases.25 Therefore, as the functional consequence of the PPP2R2B deletion is not yet known and it still could be a genetic modifier, we cannot draw a conclusion regarding the clinical relevance of this deletion and thus remains of unknown significance.
Because SNP array approaches have limitations, including low-resolution genotyping of the genome, we may have missed smaller CNVs at the exon level in SCA genes, leading to an underestimation of the occurrence of clinically relevant CNVs in the SCA genes studied. In the case of ITPR1, 3 studies used techniques (array comparative genomic hybridization and multiplex ligation-dependent probe amplification assay) able to identify smaller deletions at the exon level and showed that smaller deletions in this gene are very rare in patients with SCA.6,8,12 It is therefore unlikely that the SNP array we used missed clinically relevant smaller CNVs in ITPR1. Moreover, we suggest performing SNP array as a feasible cost-effective diagnostic screening approach to identify ITPR1 deletions in patients with SCA. However, this may not be the case for the other SCA genes, and we still may have missed small CNVs. One such example is the ∼10.8-kb deletion identified recently in SPTBN2, which we might have missed in our patients due to the limited resolution of SNP array.19 This resolution issue may be overcome in the near future as NGS has now evolved into a promising strategy to comprehensively characterize CNVs. NGS platforms can detect exon deletions, with a resolution that depends on the coverage, and can additionally define breakpoints and more accurately estimate CNVs.26,27 Therefore, to avoid missing CNVs in SCA genes, we propose using NGS platforms in the future that will increase the current diagnostic yield for SCA.
In conclusion, we systematically analyzed a cohort of patients with cerebellar ataxia for CNVs in 36 SCA genes and found that CNVs detected with the limited resolution of SNP array are a very rare cause of SCA. We found only 2 SCA genes (ITPR1 and PPP2R2B) that carried a putative clinically relevant CNV deletion. The CNV deletion we detected in ITPR1 explained the cerebellar ataxia in 2 related patients. The clinical significance of the CNV deletion in PPP2R2B remained unknown, and further studies should be performed to determine its functional consequence. Because NGS-based approaches are currently part of the routine diagnostic workflow for patients with cerebellar ataxia, we suggest adding CNV analysis of SCA genes alongside SNV analysis, at least for ITPR1, to improve the genetic diagnostics of patients with SCA.
Acknowledgment
The authors sincerely thank all the patients for their participation in this study. The authors thank Kate Mc Intyre for proofreading the manuscript.
Glossary
- BAF
B allele frequency
- CNV
copy number variant
- NGS
next-generation sequencing
- PP2A
protein phosphatase 2A
- SCA
spinocerebellar ataxia
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
Appendix. Authors
Study Funding
Rosalind Franklin Fellowship by University Medical Center of Groningen (D.S.V.). PhD grant by the GSMS of the University of Groningen (F.G.).
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/NG for full disclosures.
References
- 1.Jayadev S, Bird TD. Hereditary ataxias: overview. Genet Med. 2013;15(9):673-683. doi: 10.1038/gim.2013.28. [DOI] [PubMed] [Google Scholar]
- 2.Müller U. Spinocerebellar ataxias (SCAs) caused by common mutations. Neurogenetics. 2021;22(4):235-250. doi: 10.1007/s10048-021-00662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paulson HL. The spinocerebellar ataxias. J Neuroophthalmol. 2009;29(3):227-237. doi: 10.1097/WNO0b013e3181b416de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thapar A, Cooper M. Copy number variation: what is it and what has it told us about child psychiatric disorders? J Am Acad Child Adolesc Psychiatry. 2013;52(8):772-774. doi: 10.1016/j.jaac.2013.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Leemput J, Chandran J, Knight MA, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3(6):e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara K, Shiga A, Nozaki H, et al. Total deletion and a missense mutation of ITPR1 in Japanese SCA15 families. Neurology. 2008;71(8):547-551. doi: 10.1212/01.wnl.0000311277.71046.a0. [DOI] [PubMed] [Google Scholar]
- 7.Iwaki A, Kawano Y, Miura S, et al. Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. J Med Genet. 2008;45(1):32-35. doi: 10.1136/jmg.2007.053942. [DOI] [PubMed] [Google Scholar]
- 8.Ganesamoorthy D, Bruno DL, Schoumans J, et al. Development of a multiplex ligation-dependent probe amplification assay for diagnosis and estimation of the frequency of spinocerebellar ataxia type 15. Clin Chem. 2009;55(7):1415-1418. doi: 10.1373/clinchem.2009.124958. [DOI] [PubMed] [Google Scholar]
- 9.Di Gregorio E, Orsi L, Godani M, et al. Two Italian families with ITPR1 gene deletion presenting a broader phenotype of SCA15. Cerebellum. 2010;9(1):115-123. doi: 10.1007/s12311-009-0154-0. [DOI] [PubMed] [Google Scholar]
- 10.Novak MJ, Sweeney MG, Li A, et al. An ITPR1 gene deletion causes spinocerebellar ataxia 15/16: a genetic, clinical and radiological description. Mov Disord. 2010;25(13):2176-2182. doi: 10.1002/mds.23223. [DOI] [PubMed] [Google Scholar]
- 11.Marelli C, van de Leemput J, Johnson JO, et al. SCA15 due to large ITPR1 deletions in a cohort of 333 white families with dominant ataxia. Arch Neurol. 2011;68(5):637-643. doi: 10.1001/archneurol.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Synofzik M, Beetz C, Bauer C, et al. Spinocerebellar ataxia type 15: diagnostic assessment, frequency, and phenotypic features. J Med Genet. 2011;48(6):407-412. doi: 10.1136/jmg.2010.087023. [DOI] [PubMed] [Google Scholar]
- 13.Castrioto A, Prontera P, Di Gregorio E, et al. A novel spinocerebellar ataxia type 15 family with involuntary movements and cognitive decline. Eur J Neurol. 2011;18(10):1263-1265. doi: 10.1111/j.1468-1331.2011.03366.x. [DOI] [PubMed] [Google Scholar]
- 14.Obayashi M, Ishikawa K, Izumi Y, et al. Prevalence of inositol 1, 4, 5-triphosphate receptor type 1 gene deletion, the mutation for spinocerebellar ataxia type 15, in Japan screened by gene dosage. J Hum Genet. 2012;57(3):202-206. doi: 10.1038/jhg.2012.5. [DOI] [PubMed] [Google Scholar]
- 15.Coebergh JA, Fransen van de Putte DE, Snoeck IN, Ruivenkamp C, van Haeringen A, Smit LM. A new variable phenotype in spinocerebellar ataxia 27 (SCA 27) caused by a deletion in the FGF14 gene. Eur J Paediatr Neurol. 2014;18(3):413-415. doi: 10.1016/j.ejpn.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 16.Planes M, Rooryck C, Vuillaume ML, et al. SCA27 is a cause of early-onset ataxia and developmental delay. Eur J Paediatr Neurol. 2015;19(2):271-273. doi: 10.1016/j.ejpn.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Amado A, Blanco MO, Repáraz-Andrade A. Spinocerebellar ataxia 27: clinical phenotype of twin sisters with FGF14 deletion. Neuropediatrics. 2017;48(2):131. doi: 10.1055/s-0037-1598110. [DOI] [PubMed] [Google Scholar]
- 18.Paucar M, Lundin J, Alshammari T, et al. Broader phenotypic traits and widespread brain hypometabolism in spinocerebellar ataxia 27. J Intern Med. 2020;288(1):103-115. doi: 10.1111/joim.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romaniello R, Citterio A, Panzeri E, et al. Novel SPTBN2 gene mutation and first intragenic deletion in early onset spinocerebellar ataxia type 5. Ann Clin Transl Neurol. 2021;8(4):956-963. doi: 10.1002/acn3.51345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Araújo Lima L, Wang K. PennCNV in whole-genome sequencing data. BMC Bioinformatics. 2017;18(suppl 11):383. doi: 10.1186/s12859-017-1802-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Hearn E, Holmes SE, Calvert PC, Ross CA, Margolis RL. SCA-12: tremor with cerebellar and cortical atrophy is associated with a CAG repeat expansion. Neurology. 2001;56(3):299-303. doi: 10.1212/wnl.56.3.299. [DOI] [PubMed] [Google Scholar]
- 22.Siddique U, Choudhury S, Chatterjee K, et al. A longitudinal quantitative analysis of gait in patients with SCA-12. Clin Park Relat Disord. 2021;5:100102. doi: 10.1016/j.prdoa.2021.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sułek-Piatkowska A, Zdzienicka E, Raczyńska-Rakowicz M, et al. The occurrence of spinocerebellar ataxias caused by dynamic mutations in Polish patients. Neurol Neurochir Pol. 2010;44(3):238-245. doi: 10.1016/s0028-3843(14)60037-2. [DOI] [PubMed] [Google Scholar]
- 24.O'Hearn EE, Hwang HS, Holmes SE, et al. Neuropathology and cellular pathogenesis of spinocerebellar ataxia type 12. Mov Disord. 2015;30(13):1813-1824. doi: 10.1002/mds.26348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahit KMTH, Tarailo-Graovac M. Genetic modifiers and rare Mendelian disease. Genes (Basel). 2020;11(3):239. doi: 10.3390/genes11030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(suppl 11):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AK, Olsen MF, Lavik LAS, Vold T, Drabløs F, Sjursen W. Detecting copy number variation in next generation sequencing data from diagnostic gene panels. BMC Med Genomics. 2021;14(1):214. doi: 10.1186/s12920-021-01059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this article cannot be made publicly available as it considers patient information. To protect patient privacy, access to the data can only be made by request from the corresponding author.