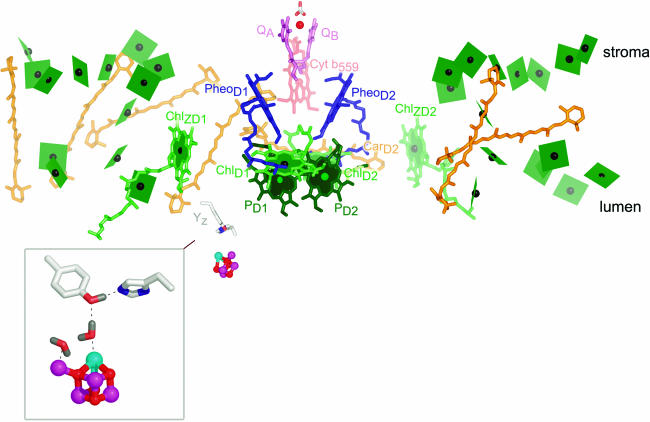
Figure 2.
Cofactors in PSII.
The cofactors in PSII, based on Protein Data Bank coordinates 1S5L, are shown. The view is perpendicular to the membrane normal. The color scheme of Ferreira et al. (2004) is retained. Chlorophylls are indicated in green, with the central Mg shown as a black sphere. The core antenna chlorophylls are distinguished from reaction center chlorophylls by the use of planes lacking phytol tails for the former versus sticks for the latter. PD1 and PD2 are in darker green to distinguish them from ChlD1 and ChlD2. Heme in cytochrome b559 is diagrammed in pale-red sticks, carotenoids in orange, pheophytin in blue, and plastoquinone in pink. The tail of plastoquinone is omitted for clarity. The metals are shown as spheres: red for iron, magenta for manganese, and cyan for calcium. The side chains of Tyr161 (YZ) and His190 near the Mn4Ca cluster are shown as gray sticks for carbon, blue for nitrogen, and red for oxygen. The inset shows an enlargement of the Mn4Ca cluster as proposed by Ferreira et al. (2004), with YZ on the top left and His190 on the bottom right. Two waters and a hydrogen on TyrZ are modeled as per Ferreira et al. (2004). Note that there is considerable ambiguity concerning the structure of the cluster (see main text). The subscripts D1 and D2 denote the polypeptide binding site of the cofactor. The polypeptide nomenclature is historical: D1 and D2 for the apoproteins that bind most of the cofactors in PSII are so named because they had been initially revealed as diffuse staining bands in thylakoid membrane preparations. CP43 and CP47 stand for chlorophyll-protein, with the numbers indicating their size as estimated from SDS-PAGE. The heme binding heterodimer is called cytochrome b559, reflecting the absorption maximum of the α band of the heme group, and other proteins are named as gene products (Psb_). PyMOL was used to generate this and other images. In all images, molecules closer to the reader are colored in a deeper shade, while those that are more distant are in a paler shade.