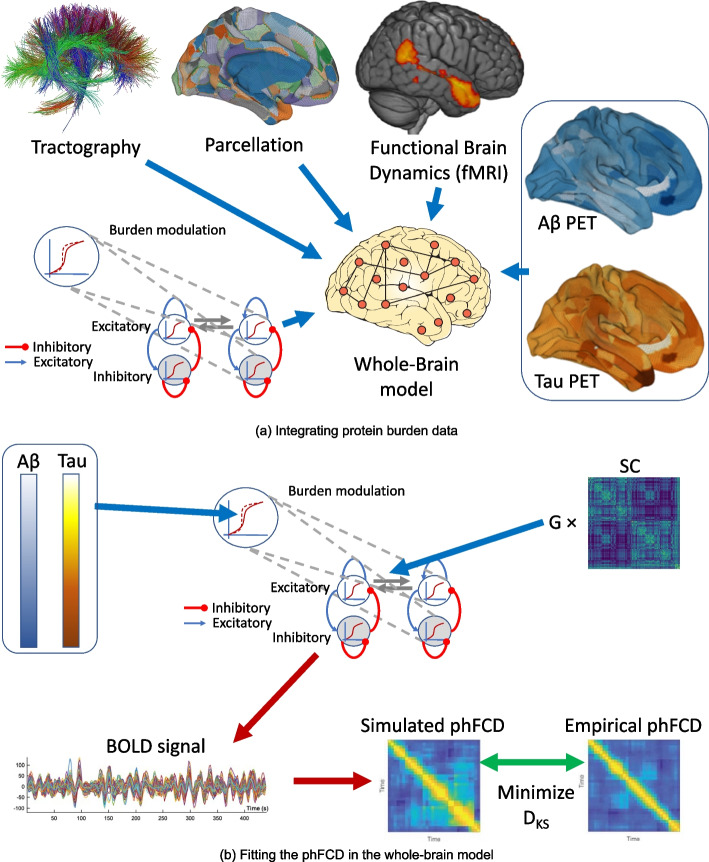
Fig. 1.
Illustrative overview of our processing pipeline. A Basic ingredients for the integration of protein burden data from structural (dMRI, top left), functional (fMRI, top right), and burden (PET, right) using the same parcellation for each neuroimaging modality (top, middle) for generating a whole-brain computational model (bottom left). Each node of the model is using a realistic underlying biophysical neuronal model including AMPA (blue connections), GABA (red), and NMDA (gray) synapses as well as neurotransmitter gain modulation of these. B Fitting the measures in the whole-brain model: First, we simulate the BOLD timeseries for each brain region in the parcellation, for each subject. These timeseries are defined by its inputs, namely a previously found global coupling constant G, an individual Structural Connectivity (SC) matrix, and the corresponding individual A and tau burdens. Subsequently, we compute a time-versus-time matrix of phase functional connectivity dynamics (phFCD). This is compared to a reference empirical phFCD extracted from the fMRI data off the same subject using the Kolmogorov-Smirnov distance (KS), , which is minimized to find the generative parameters of the model. This process is repeated for the other two measures of brain dynamics, functional connectivity (FC) and sliding-window functional connectivity dynamics (swFCD)