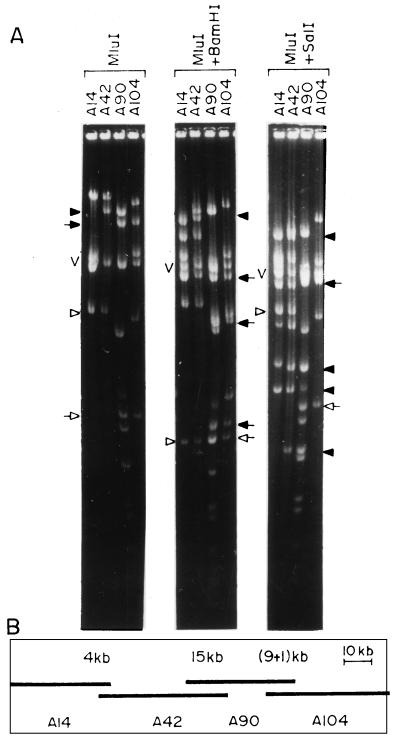
FIG. 2.
Identification of overlapping clones and contig assembly by landmark analysis. (A) MluI, MluI-plus-BamHI, and MluI-plus-SalI digestion patterns of four cosmid clones. The closed arrowhead in the MluI digest represents fragments common to cosmid clones A42 and A90, which is not cleaved by BamHI but produced four fragments (closed arrowheads) following SalI digestion. The closed and open arrows represent two fragments common to cosmid clones A90 and A104. In the MluI-BamHI double digest, both the fragments disappeared and identical new fragments appeared (closed and open arrows). In the MluI-SalI double digest, only the fragment identified by the closed arrow disappeared and identical new fragments appeared (closed arrow). The open arrowhead represents a fragment common to A42 and A14 in the MluI digest which disappeared following BamHI digestion, producing identical new fragments (open arrowhead). The MluI fragment common to A42 and A14 did not disappear upon digestion with MluI plus SalI (open arrow). V, vector DNA. (B) Assembled contig comprising four overlapping cosmids, A14, A42, A90, and A104. The extent of overlap between the clones is marked above each overlap.