Fig. 5.
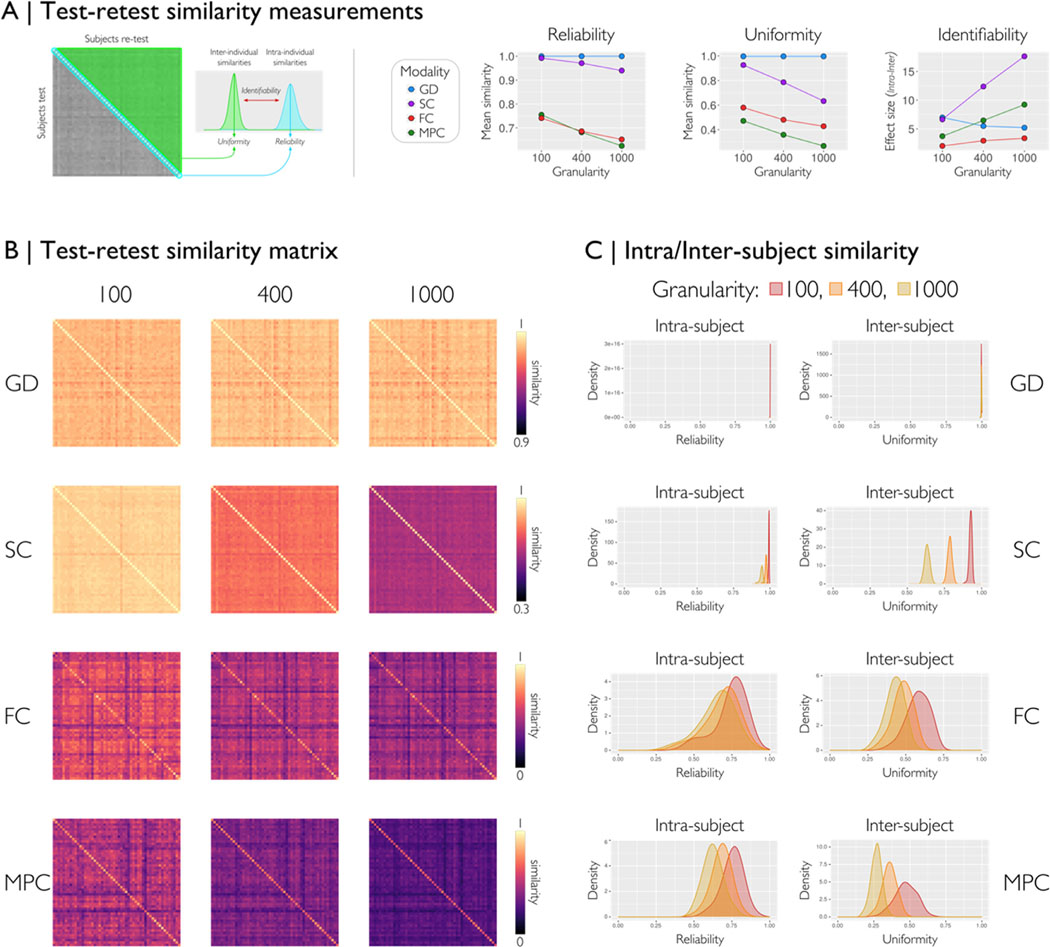
We assessed the capability of micapipe to generate reproducible outcomes in a test-retest scenario, adoping a prior framework (Seguin et al., 2022). For all modalities and three parcellations, we evaluated the similarity between matrices of two different acquisitions (53 subjects from HCP, run-1 and run-2 scans). (A) From each similarity matrix of subject-test by subject-retest, we computed three measures of similarity: reliability (intra-subject), uniformity (inter-subject), and identifiability (effect size between intra- and inter-). Reliability quantifies the mean processing consistency for an individual; uniformity quantifies the mean conformity of matrices belonging to different individuals, and identifiability quantifies how an individual can be recognized from a group based on the matrix features. The scatter plots with lines show the mean values of each similarity measure for each modality over three granularities (Schaefer-100, 400 and 1000). (B) Similarity matrices for each modality and granularity. (C) Density plots of the reliability and uniformity by modality and granularity of all subject pairs. For all feature matrices, we found higher reliability than uniformity, with excellent performance for GD and SC, and good results for FC and MPC. Overall, less granular parcellation data had higher similarity than more granular parcellation data across all modalities. GD=geodesic distance, SC=structural connectome, FC=functional connectome, and MPC=microstructural profile covariance.