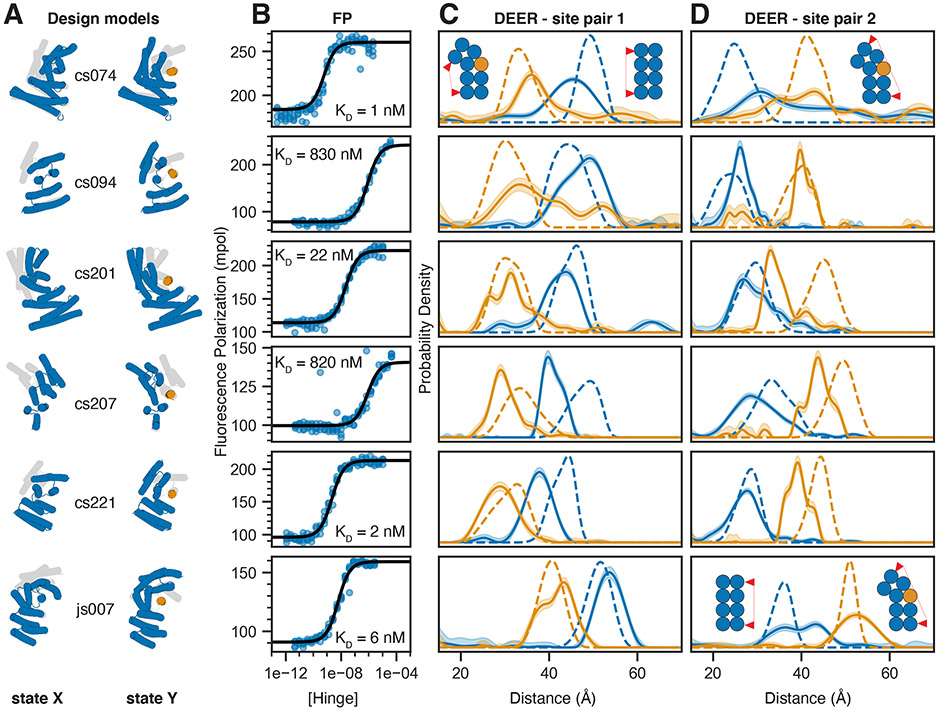
Figure 2: Experimental validation of peptide-binding hinges.
A) Design models of hinges (blue) and peptides (orange) in state X (left model) and state Y bound to the peptide (right model). Gray shades behind models in state X and Y indicate the corresponding states Y and X, respectively. B) Fluorescence Polarization (FP) titrations with a constant concentration of TAMRA-labeled peptide (0.1 nM for cs074 and cs221; 0.5 nM for cs201; 1 nM for cs094, cs207, and js007) and varying hinge concentrations. Circles represent data points from four independent measurements, lines are fits of standard binding isotherms to all data points, dissociation constants (KD) are obtained from those fits. C,D) Distance distributions between spin labels covalently attached to cysteine side chains. Solid lines are obtained from DEER experiments without (blue) or with (orange) an excess of peptide, shaded areas are 95% confidence intervals, and dashed lines are simulated based on the design models for state X (blue) or the state Y complex (orange). For each hinge two different label site pairs were tested, one in which the distance was expected to decrease with peptide binding (C) and one in which the distance was expected to increase upon peptide binding (D). Chemically synthesized peptides were used for all measurements except for cs074 site pair 1, for which sfGFP-peptide fusion was used. For design cs094, the residual state X peak in presence of the peptide can be explained by incomplete binding either due to weak binding affinity or to insufficient peptide concentration.