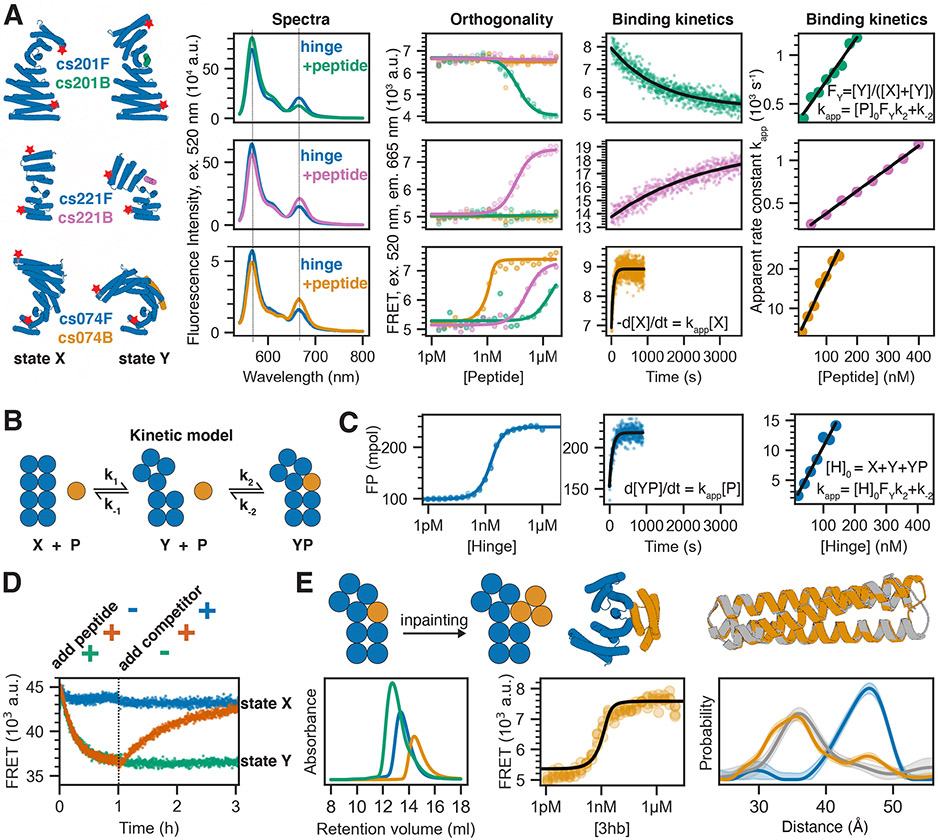
Figure 4: Quantitative analysis of conformational changes in designed hinge proteins.
A) FRET-based characterization of three extended hinges. From left to right: cylindrical representation of extended hinges (blue) and their corresponding target peptides (green: cs201B, pink: cs221B, orange: cs074B) with red stars indicating attachment sites for fluorescent dyes; fluorescence spectra (excitation at 520 nm) of labeled hinge without (blue) or with (green/pink/orange) target peptide; FRET-based binding titrations (excitation 520 nm, emission 665 nm) at 2 nM labeled hinge and varying peptide concentrations fitted with standard binding isotherms (solid lines); time course after mixing 2 nM (cs201F, cs074F) or 5 nM (cs221F) labeled hinge and 100 nM peptide fitted with a single-exponential equation (black line); apparent rate constants obtained from single-exponential kinetic fits plotted against absolute peptide concentrations (circles) and fitted with a linear equation (black line). Dotted lines in spectra indicate acceptor and donor emission peaks. B) Kinetic model describing the coupling of the conformational equilibrium to the binding equilibrium. X and Y: hinge in state X and Y, respectively; P: peptide; YP: peptide bound to hinge in state Y. k1, k−1, k2, and k−2 are the microscopic rate constants. C) FP characterization of unlabeled extended hinge cs074F. From left to right: binding titration at 0.1 nM TAMRA-labeled peptide and varying hinge concentrations; time course after mixing 2 nM TAMRA-labeled peptide and 100 nM hinge fitted with a single-exponential equation (black line); apparent rate constants obtained from single-exponential kinetic fits plotted against absolute hinge concentrations (circles) and fitted with a linear equation (black line). D) FRET-based reversibility experiment using the labeled extended hinge cs201F introduced in C). Hinge concentration is 30 nM for all traces; 1 μM peptide is added at t=0 (green/orange), 3 μM unlabeled competitor hinge is added after 1 h (blue/orange). E) Top from left to right: schematic representation of the inpainting procedure that adds two helices to the peptide cs074B yielding a three-helix bundle (3hb); cylindrical representation of 3hb_05(orange) bound to hinge cs074 (blue); overlay of design model (orange) and crystal structure (gray) of 3hb_05. Bottom from left to right: SEC traces for hinge cs074 (blue), 3hb_05 (orange), and a mixture of both (green); FRET-based titration of 2 nM extended labeled hinge cs074F and varying concentrations of 3hb_05 fitted with a standard binding isotherm (back line); Distance distributions obtained from DEER experiments as described in Figure 2 (blue: cs074, gray: cs074 + peptide cs074B, orange: cs074 + 3hb_05).