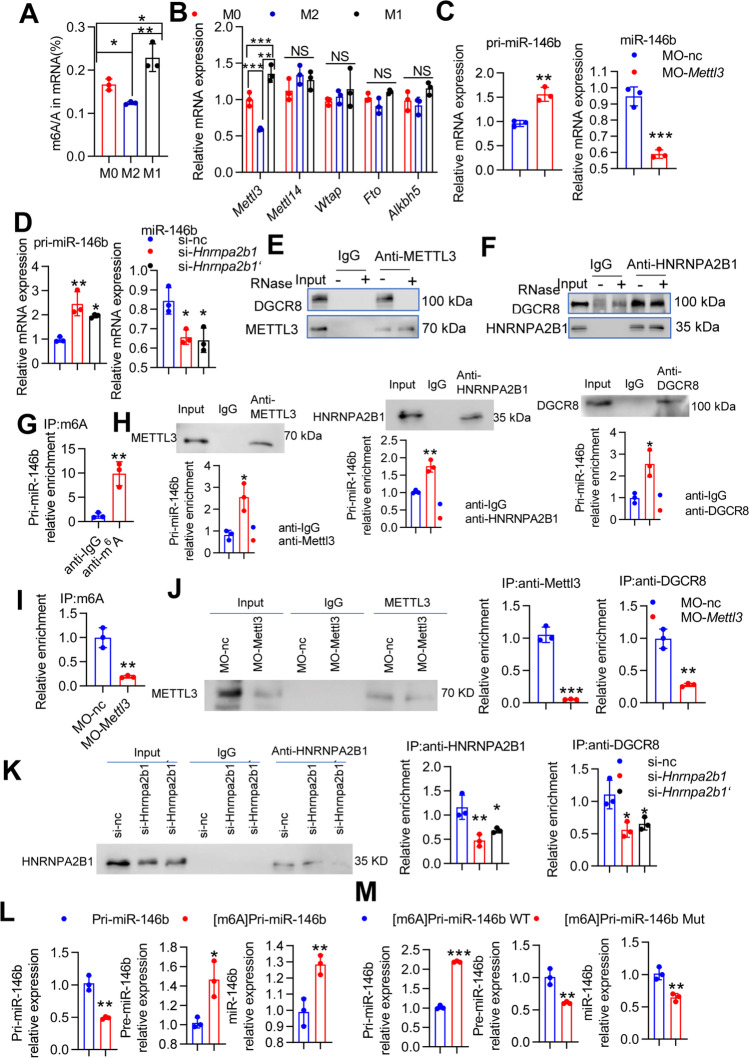
Fig. 6.
m6A modification mediates pri-miR-146b maturation in M2 macrophages. (A-B) BMDMs from WT mice were stimulated with IL-4 plus IL-10 (M2) or LPS plus IFNγ (M1), and the m6A levels of mRNAs were assessed using the EpiQuik m6A RNA Methylation Quantification Kit. Mettl3, Mettl14, Wtap, Alkbh5 and Fto mRNA levels were determined by qPCR. (C-D) BMDMs from WT mice were transfected with MO-Mettl3 or si-hnrnpa2b1 for 24 h and then stimulated with IL-4 plus IL-10 for 4 h. The mRNA levels of pri-miR-146b and miR-146b were determined by qPCR. (E) RAW264.7 cells were treated with IL-4 (20 ng/ml) plus IL-10 (10 ng/ml) for 24 h to generate M2 macrophages. Coimmunoprecipitation of the METTL3-interacting protein DGCR8 was performed. After immunoprecipitation, the samples were washed and incubated with RNase as indicated. Western blotting was used to detect METTL3 and DGCR8, and IgG served as a control for immunoprecipitation. (F) RAW264.7 cells were treated with IL-4 plus IL-10 for 24 h. Coimmunoprecipitation of the HNRNPA2B1-interacting protein DGCR8 was performed. After immunoprecipitation, the samples were washed and incubated with RNase as indicated. Western blotting was used to detect HNRNPA2B1 and DGCR8. (G) RAW264.7 cells were treated with IL-4 plus IL-10 for 24 h. Coimmunoprecipitation was performed with an anti-m6A antibody. After immunoprecipitation, the samples were washed, and pri-miR146b expression was determined by qRT-PCR. (H) RAW264.7 cells were treated with IL-4 plus IL-10 for 24 h. Western blot analysis was used to detect METTL3, HNRNPA2B1 and DGCR8 expression in the input or immunoprecipitation group, and RIP assays showed significant enrichment of pri-miR146b in RNA binding to METTL3, HNRNPA2B1 and DGCR8. (I) RAW264.7 cells were transfected with MO-Mettl3 for 24 h and then stimulated with IL-4 plus IL-10 for 4 h. Immunoprecipitation of m6A-modified RNA in control or METTL3-knockdown cells was followed by qRT-PCR to assess pri-miR146b m6A modification levels. (J) Western blot analysis was used to detect METTL3 expression in the input and immunoprecipitation group, and RIP assays showed pri-miR146b binding to METTL3 or DGCR8. (K) RAW264.7 cells were transfected with si-hnrnpa2b1 for 24 h and then stimulated with IL-4 plus IL-10 for 4 h. Western blot analysis was used to detect HNRNPA2B1 expression in the input and immunoprecipitation group, and RIP assays showed pri-miR146b binding to HNRNPA2B1 or DGCR8. (L) In vitro reaction system containing the starting materials of pri-miR-146 or [m6A]pri-miR-146 and the whole cellular lysates of 293 T cells transfected with plasmids carrying DROSHA and DGCR8. pri-miR-146b, pre-miR-146b and miR-146b mRNA levels were determined by qPCR. Pri-miR-1–1, which has no methylation site, was included as a control. (M) In vitro reaction system containing the starting materials of [m6A]pri-miR-146 WT or [m6A]pri-miR-146 Mut. pri-miR-146b, pre-miR-146b and miR-146b mRNA levels were determined by qPCR. All experiments were repeated three times independently. The data represent the mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; NS, not significant