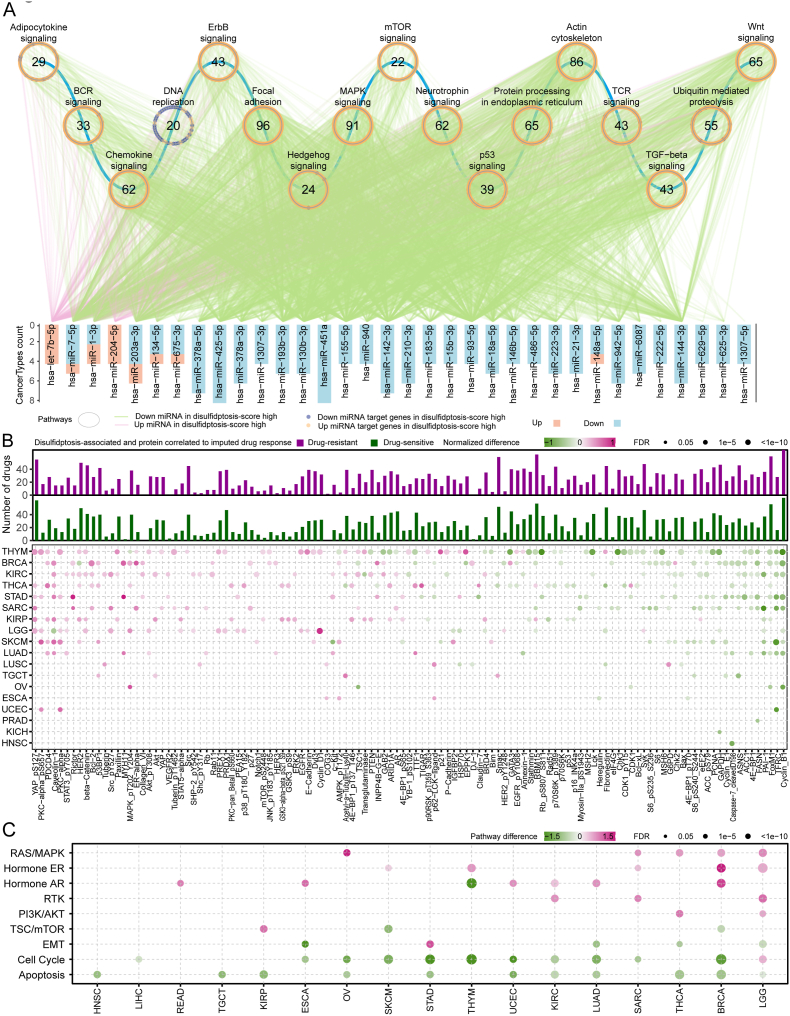
Fig. 5.
Disulfidptosis-related miRNA and protein signatures. (A) Significant Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched by mRNAs in the miRNA-target regulatory network. The magenta line indicates an upregulate of miRNA in the high disulfidptosis score group, and the green line indicates a downregulate of miRNA the high disulfidptosis score group. Yellow dots correspond to miRNA-targeted genes highly expressed in the high disulfidptosis score group, and purple dots correspond to miRNA-targeted genes highly expressed in the disulfidptosis score group. The circle represents a signaling pathway enriched with targeted genes. Statistical analysis was performed using a propensity score algorithm to identify disulfidptosis-related miRNAs and proteins; details are given in the Methods. (B) Heatmap at the bottom panel display alterations of protein or protein phosphorylation in disulfidptosis score-high samples in at least three cancer types. Color represents the degree of difference between the disulfidptosis score-high vs low groups. Statistical analysis was performed using a propensity score algorithm to identify disulfidptosis-associated proteins (details are given in the Methods). The barplot at the upper panel shows the accumulated number of drugs positively (purple) or negatively (dark green) correlated with disulfidptosis-related proteins in different cancer types. The correlation was performed by Spearman's rank correlation. (C) Altered signaling pathways based on functional proteomics data of reverse-phase protein arrays in the disulfidptosis score-high group vs. the disulfidptosis score-low group for multiple cancers. Color indicates the difference in pathway score; point size indicates FDR for pathway score. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)