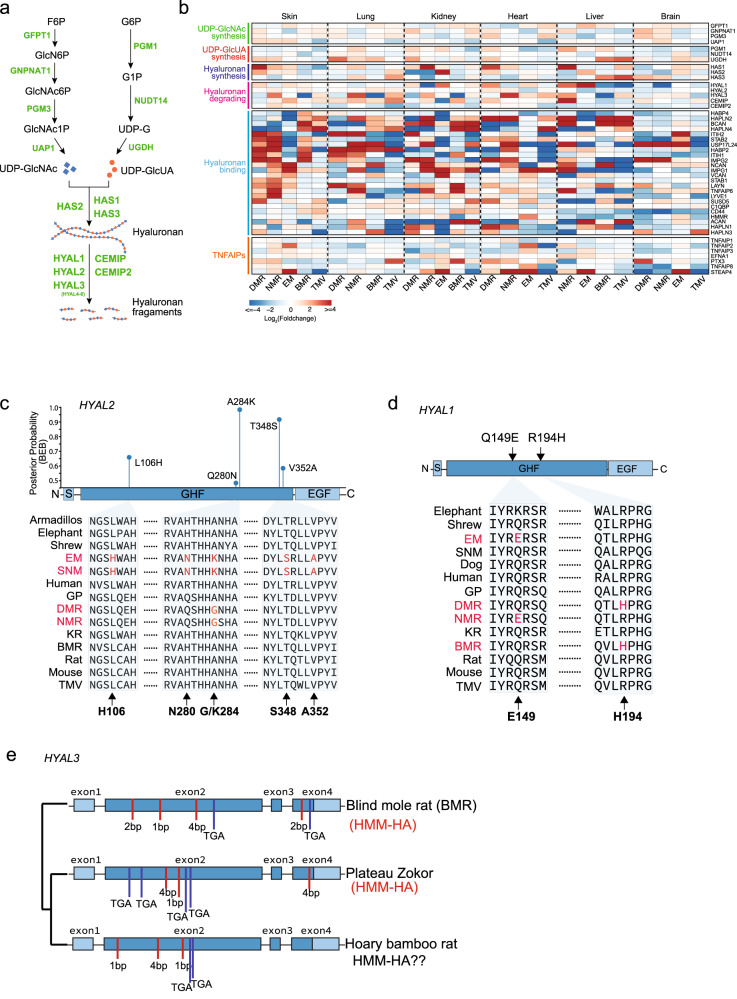
Fig. 3. Comparative analysis of gene expression and molecular evolution of genes involved in HA synthesis and degradation in subterranean species.
a HA synthesis and degradation pathway. Genes responsible for each enzymatic reaction are labeled in green. F6P Fructose 6-phosphate, GlcN6P Glucosamine-6-phosphate, GlcNAc6P N-acetyl-glucosamine 6-phosphate, GlcNAc1P N-acetyl-glucosamine 1-phosphate, UDP-GlcNAc Uridine diphosphate N-acetylglucosamine, G6P Glucose 6-phosphate, G1P Glucose 1-phosphate, UDP-G Uridine diphosphate glucose, UDP-GlcUA Uridine diphosphate glucuronic acid. b Heatmap showing the expression fold changes for HA-related genes. Genes are grouped into six sets corresponding to their function, including HA substrate metabolic pathways (UDP-GlcNAc synthesis and UDP-GlcUA synthesis), HA synthesis, HA degradation, HA binding, and TNFAIP6 related genes. The fold change values were calculated by comparing each subterranean species to their closest aboveground species. For tissues where the closest aboveground species samples are not available, rat samples were used. c Diagram on top shows the domain structure of HYAL2 and corresponding Bayes Empirical Bayes posterior probabilities (PP) of positive selection in the common ancestor of SNM and EM, with sites predicted to be under positive selection in blue (PP > 0.5). Amino acid alignment of the positively selected sites is shown below. S: Signal peptide. GHF Glycoside hydrolase family 56 (Pfam). EGF EGF-like domain. d Convergent amino acid replacements between BMR and DMR or between NMR and EM. e Diagram showing the disruptive mutations identified in GHF domain of HYAL3 of BMR and two closely related species, Bamboo rat and Plateau zokor. Red bars: indel; Blue bars: stop-codon.