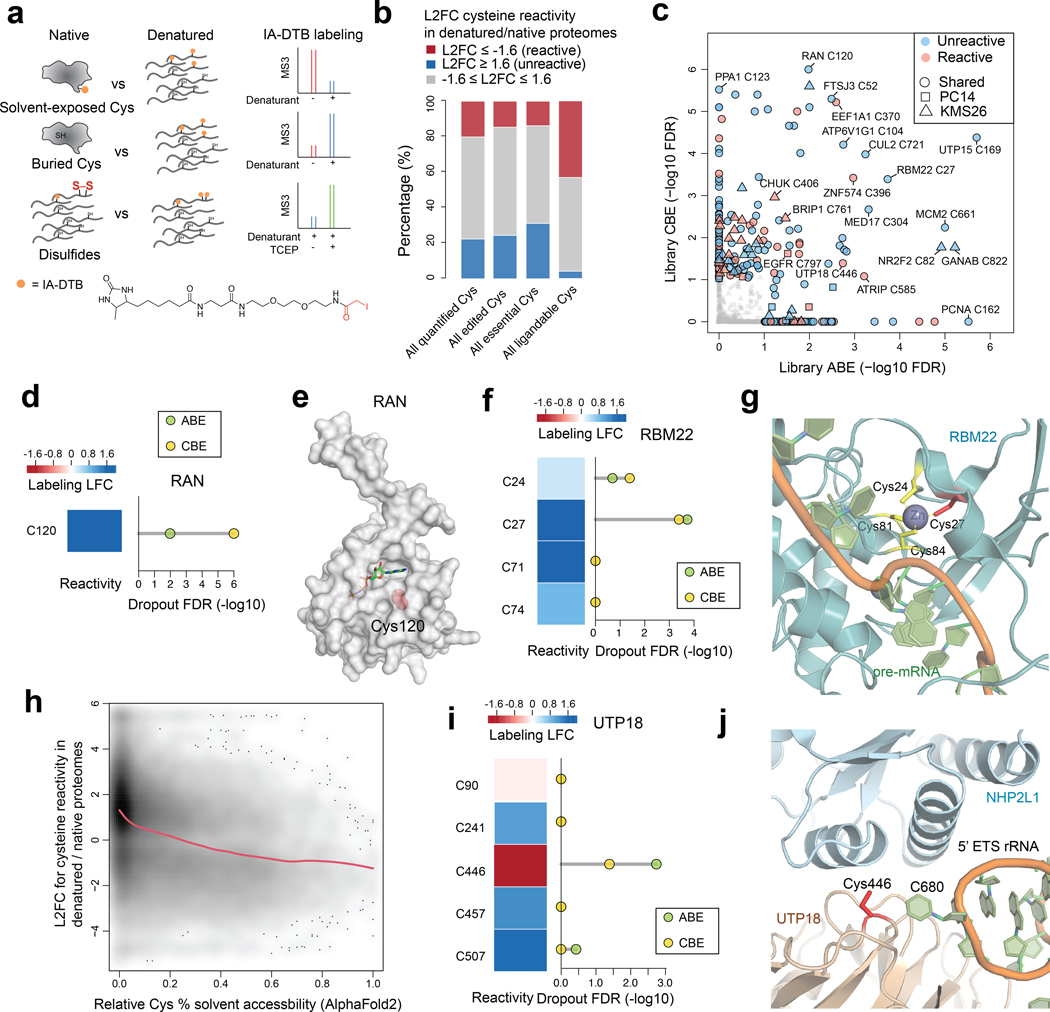
Fig. 5 |. Prioritizing essential cysteines with ligandability potential by quantitative cysteine reactivity profiling in native and denatured proteomes.
a, Schematic showing an ABPP workflow to distinguish solvent-exposed and buried cysteines based on quantitative differences in IA-DTB reactivity in native and denatured proteomes. b, Stack barplots summarizing the distribution of reactivity changes for the indicated categories of cysteines, including 1) all cysteines quantified in the ABPP experiments of native versus denatured proteomes; 2) all cysteines that were base-edited; 3) all essential cysteines (i.e., cysteines with editing-induced dropouts LFC<−0.6, FDR<10%); and 4) all ligandable cysteines using electrophilic fragments. c, Summary of editing-induced dropout values for cysteines showing substantially greater or lesser reactivity in native proteomes (designated as reactive and unreactive, respectively). Substantial reactivity changes were defined as cysteines showing denaturing/native LFC scores < −1.6 (reactive) or > 1.6 (unreactive). d, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for the edited C120 in RAN. e, Structure of RAN in complex with GNP (PDB: 5JLJ). The side chain of essential, unreactive (buried) cysteine C120 is highlighted in red. f, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in RBM22. g, Structure of RBM22 in complex with pre-mRNA as part of the human spliceosome (PDB: 5XJC). h, Density plot showing the association between predicted cysteine solvent accessibility based on AlphaFold2 protein structures and quantified cysteine reactivity values. A fitted smoothing spline is shown in red. Pearson correlation = −0.36 with two-sided p = 2.2e-16. i, The reactivity in denatured/native proteomes (left) and significance of dropout (right) for cysteines in UTP18. j, Structure of UTP18 as part of the human small subunit processome (PDB: 7MQ8). For b, c, d, f, h, i, the ABPP data of native versus denatured proteomes represent average values from 6 independent experiments. For c, d, f, i, the one-sided p values were first estimated by randomization tests of dropout data from two independent base-editing experiments and then adjusted by Benjamini-Hochberg procedure to calculate the FDR.