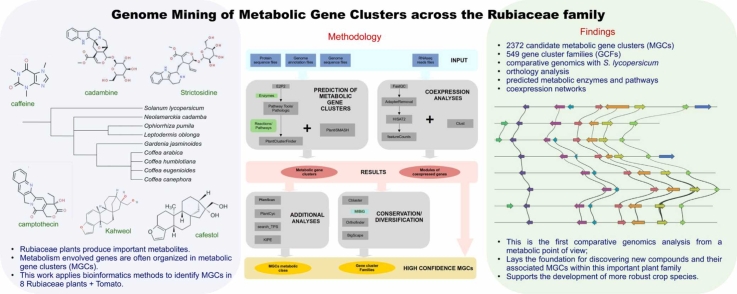
Abstract
The Rubiaceae plant family, comprising 3 subfamilies and over 13,000 species, is known for producing significant bioactive compounds such as caffeine and monoterpene indole alkaloids. Despite an increase in available genomes from the Rubiaceae family over the past decade, a systematic analysis of the metabolic gene clusters (MGCs) encoded by these genomes has been lacking. In this study, we aim to identify and analyze metabolic gene clusters within complete Rubiaceae genomes through a comparative analysis of eight species. Applying two bioinformatics pipelines, we identified 2372 candidate MGCs, organized into 549 gene cluster families (GCFs). To enhance the reliability of these findings, we developed coexpression networks and conducted orthology analyses. Using genomic data from Solanum lycopersicum (Solanaceae) for comparative purposes, we provided a detailed view of predicted metabolic enzymes, pathways, and coexpression networks. We bring some examples of MGCs and GCFs involved in biological pathways of terpenes, saccharides and alkaloids. Such insights lay the groundwork for discovering new compounds and associated MGCs within the Rubiaceae family, with potential implications in developing more robust crop species and expanding the understanding of plant metabolism. This large-scale exploration also provides a new perspective on the evolution and structure-function relationship of these clusters, offering opportunities for the highly efficient utilization of these unique metabolites. The outcome of this study contributes to a broader comprehension of the biosynthetic pathways, elucidating multiple aspects of specialized metabolism and offering innovative avenues for biotechnological applications.
Keywords: Metabolic gene cluster, Comparative genomics, Rubiaceae
Graphical Abstract
1. Introduction
Plant natural compounds are the main source of bioactives for medicinal, pharmaceutical, agricultural and industrial applications [59]. The antimalarial artemisinin, anti-cancer paclitaxel, the codeine analgesic and anti-diabetic metformin are some of the many examples of plant-derived pharmaceuticals [54], [59]. In ecosystems, plant bioactive compounds have many ecological functions, such as adaptation to the abiotic and biotic environment, defense against pests and pathogens, competition for nutrients and signaling for seed dispersal pollinators [32], [41], [47]. It is estimated that more than 200,000 known metabolites are products of plant metabolism [26]. In biosynthetic pathways, which are a series of biochemical steps that will result in a metabolite, genes involved can either be dispersed across multiple chromosomes or be organised in a physically proximate manner. Although there are many compounds, only a few have well-established metabolic pathways supported by genomic information. Over the past decade, the discovery of biosynthetic compounds has been aided by the development of genome mining and omics approaches, as reviewed by Singh et al. [51] and Zhao & Rhee [68]. Genes that compose a biosynthetic pathway can be organized as pairs, tandem arrays and biosynthetic or metabolic gene clusters [53]. A metabolic gene cluster is formed when a set of at least three genes that are of distinct evolutionary origin and are co-localized in the genome contribute to a specific metabolic pathway, ideally acting sequentially [36], [53]. The structure of an MGC often encodes enzymes responsible for creating the core metabolite and tailoring enzymes that modify this structure along with regulatory transcription factors and transporters that carry metabolites and necessary precursors. There are several examples of MGCs discovered using omics approaches in plants, as reviewed in [62] and [67]. For example, there is a gene cluster involved in the production of terpenoids in Tomato. The MGC consists of one alcohol oxidase, 5 terpene synthases, 2 cis-prenyl transferases and one functional cytochrome P450 that work together to produce mono and diterpenes in the petiole part of the leaf [34], [35]. Tohge and Fernie [58] well illustrate in their review cases where the genomic clustering of specialized metabolite genes result in the synthesis of a given compound. More recently, with the large-scale analysis of MGCs across several genomes it was possible to group putative homologous MGCs into gene cluster families (GCFs) [25]. Gene cluster families are groups of MGCs that are functionally closely related and encode the production of the same or very similar molecules [25]. This concept has been employed in genome mining for bacteria [37] and fungi [48]. Most of the studies that identify plant MGCs focus on one species, as the case of MGCs identification in tobacco [11] or selected species from distinct families [49], and more recently, MGCs discovery is starting to be incorporated more frequently in studies reporting the assembly and annotation of new plant genomes [29], [31], [64], [66].
The Rubiaceae is a plant family in the Magnoliopsida class, containing 3 subfamilies [6], more than 600 genera and 13,000 species [33]. The first genome of a Rubiaceae family plant, Coffea canephora, was published in 2014 [13]. Since then, several other genomes have been documented in the literature (Fig. 1; [7], [8], [9], [19], [22], [28], [45], [46], [61], [65], [69]. Plants from this family produce not only well-known alkaloids like caffeine, but other metabolites with great pharmacological potential, including several terpenoids [28], [2]. For example, camptothecin is a monoterpene indole alkaloid produced by Ophiorrhiza pumila that possesses antitumor activities [52].
Fig. 1.
Phylogenetic representation of the eight plant species of Rubiaceae analyzed in this study and tomato (Solanum lycopersicum) as an outgroup.
There is a small number of studies analysing the genomic basis of plant bioactive compounds synthesis in the Rubiaceae family. Some examples are the study of the crocin metabolic pathway in Gardenia jasminoides [65], caffeine in Coffea canephora [13], [40], monoterpene indole alkaloids (MIAs) in Ophorrhiza pumila [46], cadambine in Neolamarckia cadamba [69], and ursolic acid in Oldenlandia corymbosa [22].
For some Rubiaceae species, i.e. Ophiorrhiza pumilla and Neolamarckia cadamba, genome analysis included the prediction of MGCs. In O. pumilla, it was found specific clusters with highly coexpressed genes, indicating their possible role in MIA biosynthesis [46]; however, a comparative family-wide analysis is still lacking.
The study advances the field of computational plant genomics by conducting a pioneering detailed comparative analysis of metabolic gene clusters (MGCs) and cluster families in a plant family, using the Rubiaceae family as a case study. By incorporating methods such as orthology assessment, gene family expansions, coexpression analysis, and a comparative analysis of metabolic gene clusters (MGCs), this investigation will facilitate the priorization of previously unknown pathways to understand the synthesis of bioactive compounds in plants. This pioneering comparative genomics research within the Rubiaceae family seeks to lay a foundational framework for the identification of new compounds and their corresponding MGCs. The potential benefits of uncovering these MGCs are manifold, including the improvement of crop resilience and the exploration of novel bioactive substances, which could have profound implications for applications in both agricultural and medicinal contexts.
2. Materials and methods
2.1. Genomic and annotation data
For our analysis, we considered Rubiaceae genomes with high-quality assemblies (chromosome-level sequencing and a BUSCO score of over 97%) with publicly available deduced proteomes and GFF-formatted genome coordinate files (Table 1; Supplementary File 1). By December 1st, 2022, eight species from three subfamilies met these criteria: Coffea arabica, Coffea canephora, Coffea eugenioides, Coffea humblotiana, Gardenia jasminoides, Leptodermis oblonga, Ophiorrhiza pumila and Neolamarckia cadamba. Table 1 summarizes these sources. We adopted Solanum lycopersicum, a Solanaceae, as an outgroup for our comparative genomics analysis, as it is phylogenetically closer to Rubiaceae than Arabidopsis thaliana (Brassicaceae), providing a more relevant comparison, with previous predictions of MGCs [35], [17], [70]. The genome sequence and annotation files from Solanum lycopersicum release SL4.0 [21] were downloaded from https://solgenomics.net/organism/Solanum_lycopersicum/genome/.
Table 1.
Species used in the present study.
| Species | Subfamily | Assembly | Authors | Source |
|---|---|---|---|---|
| Coffea arabica | Ixoroidea | Cara_1.0 | Johns Hopkins University | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_003713225.1 |
| Coffea canephora | Ixoroidea | AUK_PRJEB4211_v1 | [13] | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_900059795.1/ |
| Coffea eugenioides | Ixoroidea | Ceug_1.0 | Johns Hopkins University | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_003713205.1/ |
| Coffea humblotiana | Ixoroidea | release 1.0 | [45] | https://solgenomics.net/organism/Coffea_humblotiana/genome |
| Gardenia jasminoides | Ixoroidea | release 1.0 | [65] | https://genomevolution.org/coge/api/v1/genomes/62692/sequence |
| Leptodermis oblonga | Rubioideae | release 1.0 | [19] | https://www.ncbi.nlm.nih.gov/data-hub/genome/GCA_016801395.1/ |
| Ophiorrhiza pumila | Rubioideae | release 1.0 | [46] | https://pumila.kazusa.or.jp/ |
| Neolamarckia cadamba | Cinchonoideae | release 1.0 | [69] | https://figshare.com/s/ed20e0e82a4e7474396b |
2.2. Identification of metabolic gene clusters (MGCs) through genome mining
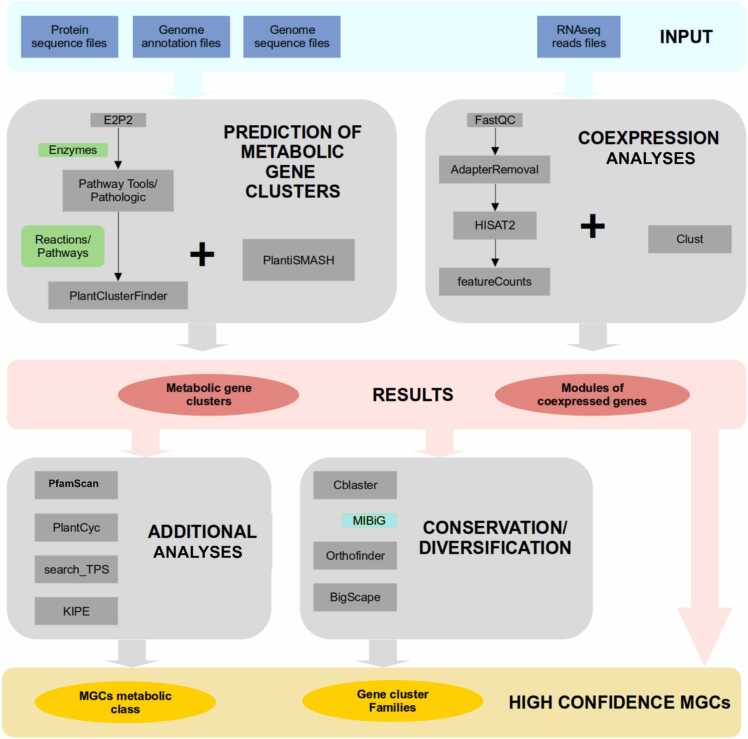
Our goal was to identify metabolic gene clusters using two genome mining tool approaches (PlantClusterFinder and PlantiSMASH), compare the results, and apply criteria to select high confidence MGCs (Fig. 2). To acquire a set of high-confidence MGCs, we consider results only from genomes with defined chromosomes.
Fig. 2.
Overview of the pipeline to predict metabolic gene clusters.
We used E2P2 v4.0 [20] and annotated protein sequences to identify enzymes associated with plant metabolic pathways and then used Pathway Tools v. 26 [23] and the PathoLogic software with default settings to generate metabolic pathway databases. These predicted pathways were manually filtered to only include those present in plants. To predict metabolic gene clusters, we modified a method based on previous studies by Schläpfer et al. [49] and Chen et al. [11] (see Fig. 2). The output file from E2P2 was used with Pathway Tools to create species-specific metabolic pathway databases. These databases were then exported and inputted into PlantClusterFinder (PCF) version 1.3 (https://github.com/carnegie/PlantClusterFinder, [49], which identifies groups of metabolic genes located contiguously on the same scaffold using sliding window searching. Default parameters were used for PlantClusterFinder. Finally, we used PfamScan v. 1.6 (https://github.com/gpertea/gsrc/blob/master/scripts/pfam_scan.pl; [15] to determine protein domains for all genes identified by PlantClusterFinder.
We also used PlantiSMASH v. 1.0, a computational pipeline that predicts plant MGCs using specific HMM profiles [24], to identify MGCs. We input the genome sequences and annotation files in GFF3 format and applied the dynamic cutoff parameter for analysis.
2.3. Clustering and evolutionary analysis of metabolic gene clusters in Rubiaceae
After identifying MGCs in Rubiaceae plants, we aimed to determine if they showed evolutionary conservation. To achieve this, first we compared the genomes of the study using Orthofinder v. 2.3.8 [16] with default parameters to infer orthology and Orthovenn3 [56] with default parameters to infer gene families expansions and contractions and synteny analysis. We then grouped the MGCs into gene cluster families, identified protein domains of predicted metabolic enzymes, and checked for orthology relationships. To classify the high-confidence MGCs into families, we utilized the Biosynthetic Gene Similarity Clustering and Prospecting Engine (BiG-SCAPE) v. 1.1.0 [38]. We applied the "mix" and "no-classify" parameters and set a cutoff value of 1.0 as a raw distance. Chae et al. [10] proposed a system to classify MGCs into 13 primary functional classes. Later, Schläpfer et al. [49] applied this system to classify MGCs detected with the PlantClusterFinder tool. In our study, we used the same strategy to classify the MGCs that we identified using the PCF tool (refer to Fig. 3A for more details).
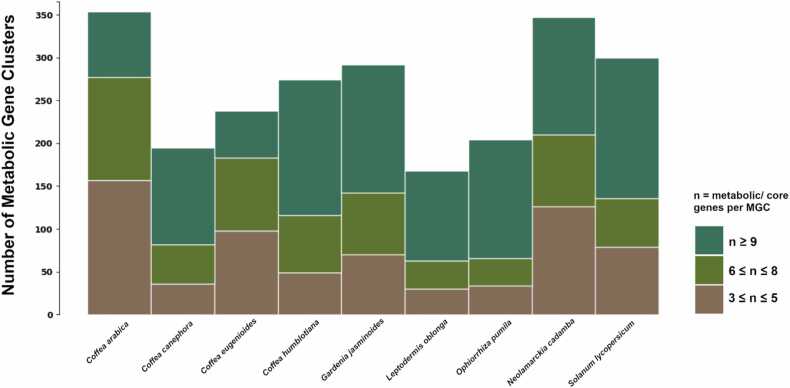
Fig. 3.
Number of all predicted metabolic gene clusters of different sizes (number of clustered metabolic genes) across 8 plants from the Rubiaceae family and one Solanaceae species.
2.4. RNA-Seq data and coexpression analysis
To determine if the predicted MGC genes were coexpressed, we constructed coexpression networks using transcriptome data for six Rubiaceae species: Coffea arabica, Coffea canephora, Coffea eugenioides, Gardenia jasminoides, Neolamarckia cadamba and Ophiorrhiza pumila. For this, we used 65 libraries from 4 RNA-Seq experiments available in the European Nucleotide Archives (ENA) and the National Genomics Data Center (NGDC). In Table 2, we detail conditions of these experiments:
Table 2.
Information of the RNA-Seq experiments used in this study.
| Project ID | Source | Experiment description | Author |
|---|---|---|---|
| PRJEB32533 | ENA | Transcriptome of seeds in three developmental stages from Coffea arabica, Coffea canephora and Coffea eugenioides | [55] |
| PRJNA352919 | ENA | Transcriptome of Ophiorrhiza pumila hairy roots | [60] |
| PRJCA003540 | NGDC | Transcriptome of Neolamarckia cadamba roots under aluminum stress | [12] |
| PRJNA688705 | ENA | Transcriptome of Gardenia jasminoides fruits in two developmental stages. | [39] |
For Coffea arabica, Coffea canephora, and Coffea eugenioides, we performed coexpression analysis using RNA-Seq of seeds at three developmental stages [55]. The study generated 27 RNA-Seq libraries (3 species, 3 replicates, 3 seed stages). Seed stages corresponded to the following phenological phases: ST5 (seeds from green fruits, peak of reserve deposition and start of endosperm hardening), ST6 (seeds during fruit veraison), and ST7 (seeds from mature cherry fruits with red pericarp). The RNA-Seq experiments in the Gardenia jasminoides dataset [39] used peel and sarcocarp samples from both green and red fruits, collected in triplicates. The study resulted in 12 RNA-Seq libraries, which were grouped into four tissues for coexpression analysis: GFS (Sarcocarps of green fruits), GFP (Peels of green fruits), FS (Sarcocarps of red fruits) and FP (Peels of red fruits). In the Ophiorrhiza pumila dataset [60], the expression of two ERF transcription factors was suppressed in hairy roots through RNA interference. The study generated 6 RNA-Seq libraries (3 conditions, 2 replicates each), which were divided into the following codes for coexpression analysis: Gusi (Hairy roots transformed with GUS); ERF1i (Hairy roots with suppressed OpERF1); ERF2i (Hairy roots with suppressed OpERF2). In the Neolamarckia cadambia RNA-Seq experiment [12], roots were treated with 400 μM +Al for 1, 3, and 7 days, while controls were grown without Al3 + . The experiment made in 20 RNA-Seq libraries, each defined by one of four time sets and two conditions: untreated roots at 0, 1, 3, and 7 days old (AL0, AL1, AL3, AL7) and treated roots at 1, 3, and 7 days old (AL1t, AL3t, AL7t). We analyzed the RNA-Seq raw data using FastQC v0.11.8 tool [63] and removed low quality reads and adapters with the AdapterRemoval v2.3.0 software [50]. Then, we mapped the data against the respective genome using HISAT2 v2.2.0 [27] with default parameters. Finally, we used the featureCounts v2.0.0 tool [30] to count and normalize the transcripts.
We carried out a coexpression analysis with the Clust v1.12.0 tool [1] using the raw count data from the RNA-Seq experiments (as listed in Table 2). The parameters used were k-means clustering method, tightness weight of 1.0, and Q3s outliers threshold of 2.0. For a cluster to be considered among those with coexpressed genes, at least three biosynthetic genes should be in the same coexpression module.
3. Results and discussion
3.1. Genome-wide prediction of metabolic gene clusters in the Rubiaceae family
The surge in large-scale transcriptomic and genomic datasets has opened new dimensions in plant comparative genomics. This work demonstrates the application of omics techniques and bioinformatics tools for discovering metabolic gene clusters. Our analysis of MGCs across eight Rubiaceae species plus Solanum lycopersicum allowed us to predict a total of 2372 metabolic gene clusters using two pipelines. In Fig. 2, we show the distribution of these genes among clusters and species.
Using the PlantClusterFinder pipeline, we identified a total of 1931 metabolic gene clusters containing 31,392 genes, with detailed results in Table 3 and supplementary Table S1. We identified an average of 214 MGCs per species, with the lowest number occurring in L. oblonga (118 MGCs) and the highest number occurring in N. cadamba (295 MGCs). The predicted MGCs ranged from 5 to 2551 kb with an average size of 178 kb. The average number of genes per MGC was 17. A total of 22,556 genes had an attributed E.C. number and 22,713 had at least one attributed reaction number. In our analysis, we identified 1069 genes within metabolic gene clusters that were also predicted in the O. pumila genome assembly [46] using PlantClusterFinder.
Table 3.
Overview of results from the PlantClusterFinder pipeline.
| Species | MGCs | Average nº of genes |
|---|---|---|
| Solanum lycopersicum | 255 | 17 |
| Neolamarckia cadamba | 295 | 14 |
| Ophiorrhiza pumila | 162 | 23 |
| Leptodermis oblonga | 118 | 26 |
| Gardenia jasminoides | 238 | 17 |
| Coffea humblotiana | 223 | 19 |
| Coffea canephora | 149 | 21 |
| Coffea eugenioides | 200 | 8 |
| Coffea arabica | 291 | 8 |
Using the PlantiSMASH pipeline we predicted 441 MGCs, which contained 5776 genes (Table 4; supplementary Table S2). On average, 49 MGCs were predicted per species, with the lowest number occurring in C. eugenioides (38 MGCs) and the highest occurring in C. arabica (63 MGCs). The predicted MGCs ranged from 18 to 960 kb, with an average size of 175 kb. The average number of genes per MGC was 13. The authors from the genome assembly of N. cadamba study (Zhao et al., 2021) also predicted MGCs using PlantiSMASH and 622 genes identified in MGCs by their study were identified in our analysis.
Table 4.
Overview of results from the PlantiSMASH pipeline.
| Species | Number of Predicted MGC’s | Average nº of genes |
|---|---|---|
| S. lycopersicum | 45 | 11 |
| N. cadamba | 52 | 10 |
| O. pumila | 42 | 16 |
| L. oblonga | 50 | 15 |
| G. jasminoides | 54 | 12 |
| C. humblotiana | 51 | 13 |
| C. canephora | 46 | 11 |
| C. arabica | 63 | 12 |
| C. eugenioides | 38 | 12 |
These pipelines are based on different methodologies and algorithms, leading to substantial discrepancies in the MGCs they predicted. PlantClusterFinder identified 41.7% of the MGCs that were predicted by PlantiSMASH, but only 7.7% of MGCs detected by PlantClusterFinder were detected again by PlantiSMASH. These differences were previously reported [49], [11] and underscore the importance of considering multiple methods in MGC discovery and the inherent complexities in these types of analyses.
We compared the identified MGCs found in Rubiaceae species were homologous to MGCs from other plants from the curated database within the "Minimum Information about Biosynthetic Gene clusters'' (MiBIG) repository [57]. Using this approach, we successfully recovered all MGCs from S. lycopersicum, indicating the effectiveness of our methodological approach. Although this repository contains 43 verified MGCs for Viridiplantae, none belong to the Rubiaceae family. To identify any plant MGCs in the MiBIG repository that could share similarities with MGCs in Rubiaceae species, we employed the cblaster tool (version 1.3.16; [18]. Out of the 43 plant MGCs in MiBIG, 23 had partial matches to MGCs from Rubiaceae species (Supplementary File 1), with low similarity (identity below 40%). Among the partially identified cases, 13 involve conserved TPS-CYP gene pairs, which are a common structure in clusters related to terpenoid metabolism [4], [5], [53]. The fact that no MGCs from other plant species were conserved in the Rubiaceae underscores the unique biosynthetic diversity identified in the genomes of this family.
3.2. Classification of Rubiaceae MGCs
In our study, we used a strategy based on a system proposed by Chae et al. [10] to classify the MGCs that we identified using the PCF tool (refer to Fig. 3A for more details). Each predicted enzyme is then designated a 'signature' or 'tailoring' classification. Out of the total MGCs we initially identified, we found that 175 of them (9%) included both 'signature' and 'tailoring' enzymes.
The PlantiSMASH pipeline attributes a biochemical class to each predicted MGC in saccharides, terpenes, alkaloids, lignans, polyketides, putative or mixed. This classification follows a criteria based on the number of core and accessory genes identified with specific pHMMs within a MGC [24]. In other MGC predictions with PlantiSMASH (N. cadamba - Zhao et al., 2021; tobacco - [44], the most identified classes were also saccharides and terpenes. In our analysis, the most frequently occurring biochemical class was saccharides (Fig. 3), with a total of 152 clusters identified, averaging 16.8 per species. The class with the lowest number of clusters was polyketides, with a total of 20 and an average of 2.2 per species. We identified 39 clusters as hybrids and the metabolic class was undetermined for 103 clusters (Fig. 3B).
Given that the selected methods employ different techniques for predicting Metabolic Gene Clusters (MGCs), we utilized an integrative analysis to consolidate the results. The PlantiSMASH tool predicts MGCs using profile Hidden Markov Models (pHMMs), so to harmonize this with the PlantClusterFinder predictions, we performed a search for protein domains in each gene within an MGC. This was executed using PfamScan (please refer to supplementary Table S3 for more details).
The protein domain family for cytochrome P450 (PF00067) was the most frequently detected, followed by the UDP-glucuronosyl and UDP-glucosyl transferase (PF00201), as well as the 2OG-Fe(II) oxygenase superfamily (PF03171).
In addition to the protein domain search, we performed a search for Metacyc plant pathways for each gene within a Metabolic Gene Cluster (MGC) that was predicted by PlantiSMASH. We chose to do this because the PlantClusterFinder (PCF) pipeline employs this methodology (see supplementary Table S3 for additional information). The most frequently identified pathway was the Secologanin and Strictosidine biosynthesis pathway (PWY-5290). Following closely were the Sesaminol Glucoside/lignan biosynthesis pathway (PWY-7139), the Quercetin Glucoside/flavonoid biosynthesis (PWY-7129), and the flavonoid biosynthesis pathway (PWY1F-FLAVSYN).
To predict and identify terpene synthases and enzymes involved in flavonoid biosynthesis within Metabolic Gene Clusters (MGCs), we employed two specialized tools: search_TPS (version 1.0; [14] and KIPEs (version 0.35; [43]. After conducting a thorough analysis, we identified several MGCs with distinct types of synthases: 50 MGCs contained monoterpene synthases, 30 displayed diterpene synthases, and 75 had sesquiterpene synthases. Furthermore, we found 199 MGCs that included genes related to flavonoid metabolism. Comprehensive details of these findings are provided in Supplementary Table S3.
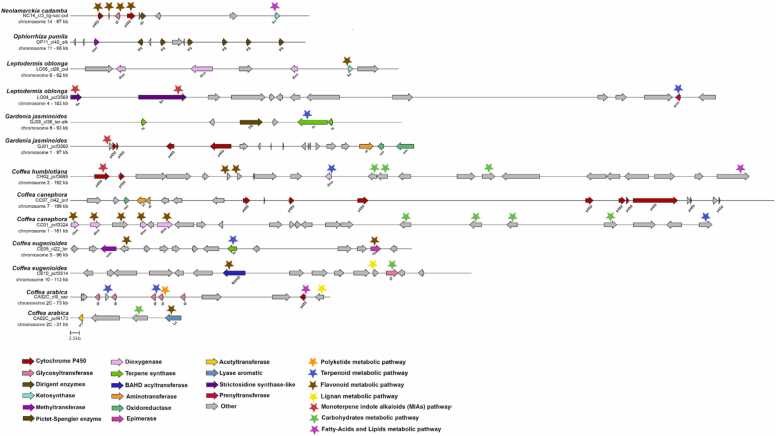
The MGC predictions unveiled diverse and often complex structures. In Fig. 5, we show examples of MGCs conserved among genomes. In terms of their functional classification, both pipelines detected a high number of saccharide and terpene MGCs, with saccharides being the most prevalent biochemical class identified.
Fig. 4.
- Percentual distribution of metabolic domains in the MGCs predicted by PCF (A). Overview of MGCs predicted with the PlantiSMASH pipeline and classified into biochemical classes (B).
Fig. 5.
- Example candidate MGCs identified in this study. The examples cover a diverse range of enzymatic classes (arrows) and predicted metabolic pathways (stars).
3.3. Conservation and diversification of metabolic gene clusters in Rubiaceae
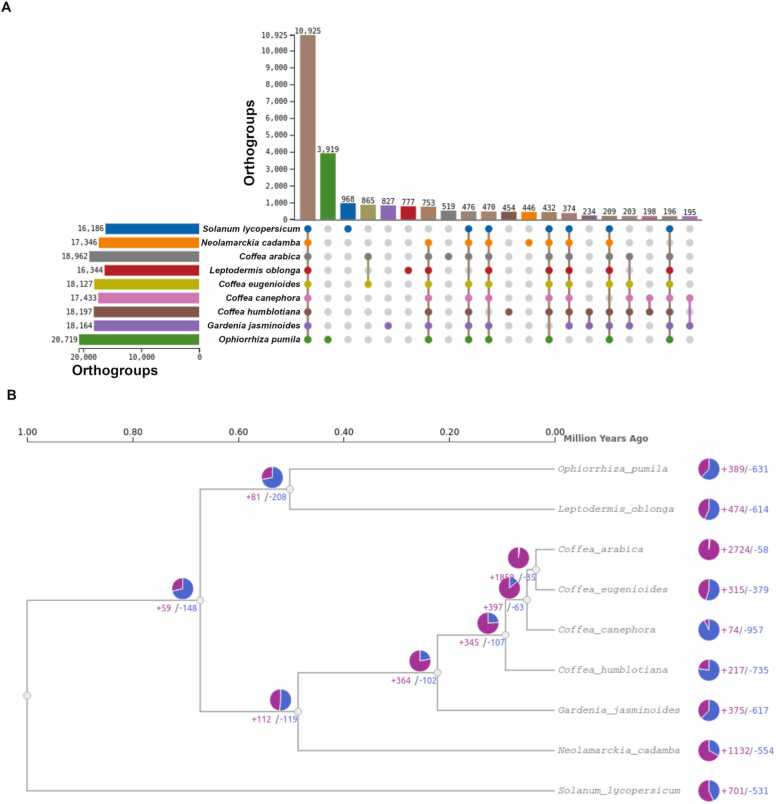
We performed a comparative genomic analysis with all plants of the study to assess conservation and diversification of metabolic gene clusters in Rubiaceae. With an orthology analysis we detected a total of 30,170 orthogroups (supplementary Table S12). A total of 10,925 orthogroups containing all species and 8152 species-specific orthogroups were identified (Fig. 6A). All nine species had species-specific orthogroups.
Fig. 6.
- Gene family contraction and expansion analysis of eight species from Rubiaceae family plus Solanum lycopersicum. (A) The UpSet table displays the count of orthogroups for each species, along with the count of unique orthogroups and the count of shared orthogroups among different species. (B) A pie chart was utilized to visualize gene families with altered gene numbers, representing the expanded gene families (depicted in purple) and contracted gene families (depicted in blue).
To investigate gene content changes, we examined the rates and direction of changes in orthogroup size among each of the species. Across the Rubiaceae phylogeny, most species have higher numbers of orthogroup contractions than expansions, except for N. cadamba, C. arabica and C. eugenioides (Fig. 6B). Orthogroups in the C. arabica genome exhibit the highest number of expansions and contractions followed by N. cadamba.
Synteny analysis among the nine species identified the biggest collinearity between C. arabica and C. eugenioides with 50932 (72.08%) collinear gene pairs. The smallest collinearity was identified between C. canephora and N. cadamba with 3075 (3.23%) collinear gene pairs.
All genes predicted in MGCs were distributed in 3121 orthogroups (supplementary Table S4).
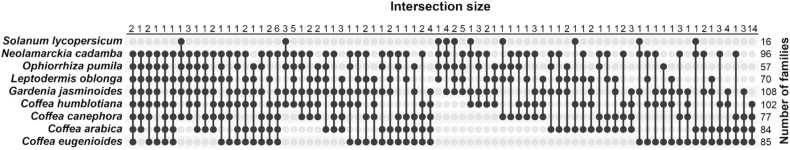
In order to track the conservation within Rubiaceae MGCs, we constructed a similarity network of MGCs and identified a total of 549 gene cluster families (GCFs) (Fig. 5; supplementary Table S5). The average number of MGCs per family was 4 and the maximum number of MGCs in a family was 16. Fig. 5 summarizes families that were found in at least four species, with the most conserved MGCs.
The results of the orthology analysis were used to validate the gene cluster families prediction - since it would be expected that genes in a given GCF would be in the same orthogroups. Of the total 549 predicted GCFs, 179 were formed by a single MGC per species (or 2 in the case of the tetraploid Arabica coffee).
It is well known that MGCs of the same family can evolve to produce different molecules, through neofunctionalization of genes or by gene recruitment or loss [42]. Thus in most of the cases of Rubiaceae MGCs, the homologous genes are distributed in different genomic positions or forming different MGCs across the plants. We observed an example of this by comparing our results with the genome assembly of Gardenia jasminoides study [65]. Xu et al. describe syntenic regions between G. jasminoides and Coffea canephora containing crocin biosynthetic genes, in which specific duplications of the genome can explain the synthesis of this compound in Gardenia and the absence in Coffea. One of these syntenic regions contains a tandem array of genes encoding the UDP-glycosyltransferase family (UGT) (PF00201.21) in Gardenia, on chromosome 9, which has an orthologous relationship with UGTs identified on chromosome 2 of C. canephora [65]. We identified a MGC in the chromosome 2 of C. canephora, CMG CC02_cl15_sac, predicted to be involved in saccharide biosynthesis, which corresponds to this region analyzed by Xu et al. [65]. C. canephora MGC was identified in our analysis because, in addition to UGTs, this region of the genome has a gene from the cytochrome P450 family (PFAM domain PF00067.25) that is not present in the homologous region of chromosome 9 in G. jasminoides. This data suggests a genomic diversification that led to a metabolic diversification between these two species in this homologous region of the genome.
We highlight two cases of MGCs that conserve core biosynthetic genes across more than five species, with shared orthogroups (Fig. 7, Fig. 8). As we used Solanum lycopersicum as an outgroup for comparative genomics analysis, we observed an example of a putative conserved MGC involved in the metabolism of saccharides. In the gene cluster family FAM-1539 we observed that both the core and accessory genes of MGCs from six Rubiaceae plants demonstrate a degree of conservation with S. lycopersicum, indicating their potential importance in the production process of a compound predicted as saccharide. The example of the family FAM-1539, which has retained MGCs in seven species: N. cadamba, O. pumila, L. oblonga, C. canephora, C. arabica, C. eugenioides, and S. lycopersicum. Each species carries one MGC from this GCF, with the exception of C. arabica which has two. These MGCs are linked to saccharide metabolism.
Fig. 9.
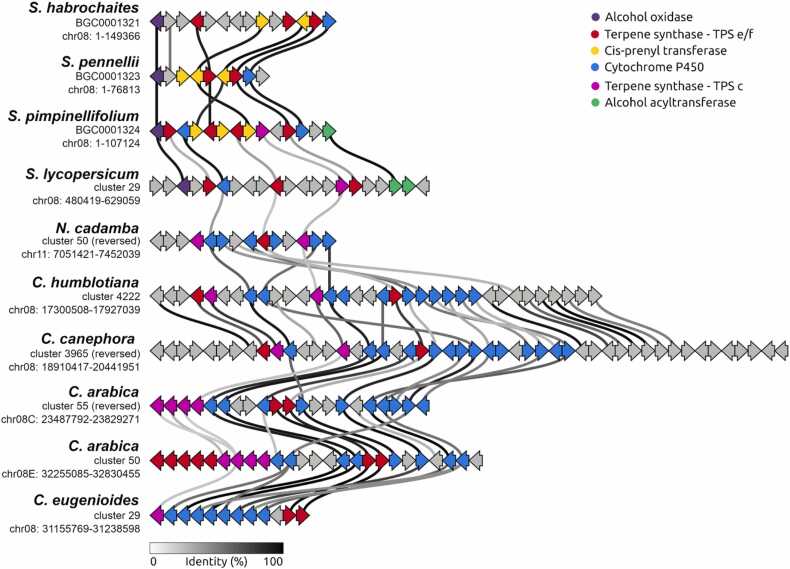
- Gene cluster family FAM-1569 with the tomato lycosantalonol MGC conserved in wild species of tomato. This MGC is partially conserved in five Rubiaceae species. This representation was plotted disregarding the scale factor due to the large difference between the sizes of the represented MGCs.
Fig. 7.
- Gene cluster families presence in four or more species across Rubiaceae and tomato.
Fig. 8.
- Gene cluster family FAM-1539 with conserved MGCs in S. lycopersicum, N. cadamba, O. pumila, L. oblonga, C. canephora, C. arabica and C. eugenioides. Each arrow represents a gene. The width of the links connecting genes represents the percentity of identity.
MGCs from FAM-1539 comprises three core biosynthetic genes (a Glycosyltransferase, a Squalene epoxidase, and an Aminotransferase) and seven accessory genes, as displayed in Fig. 6. Notably, we observed that both the core and accessory genes demonstrate a degree of conservation, indicating their potential importance in the compound production process for this specific set of MGCs. In the case of tomato (S. lycopersicum), the glycosyltransferase gene is a cellulose synthase (Solyc04g077470, domain PF13632.9), an enzyme usually involved in the synthesis of matrix polysaccharides such as xyloglucan. The tomato aminotransferase is an ACC synthase paralog (Solyc04g077410, domain PF00155.24), a key enzyme implicated in the synthesis of ethylene.
We analyzed the expression of orthologs of two core genes of the tomato MGC in this gene cluster family (cellulose synthase and ACC synthase) and observed that they are not the most expressed of their respective gene families. We also observed that such orthologs are not coexpressed, however, this conservation suggests that the synthetic processes of polysaccharides and hormones are physically linked in the genome of several species of Rubiaceae. Future functional studies interrupting or overexpressing these genes would help in the final understanding of the function of this MGC.
The second highlighted example involves the partial preservation of a tomato terpenoid MGC in Rubiaceae plants (FAM-1569). This cluster of genes in tomato (Solanum lycopersicum) has been found to be involved in the synthesis of mono, sesqui and diterpenes. The tomato MGC contain five complete terpene synthase genes (TPS18, TPS19, TPS20, TPS21, and TPS41), two complete cis-prenyl transferases (CPTs), a cytochrome P450s, an aldehyde oxidase, and three alcohol acyl transferase genes [34]. This cluster evolved via gene duplication, divergence, alterations in substrate specificity, and acquisition of cis-prenyl transferase genes in wild tomato species, such as Solanum habrochaites, S. pennellii, and S. pimpinellifolium. FAM-1569 (Fig. 7) includes, besides tomato, MGCs from N. cadamba, C. humblotiana, C. canephora, C. arabica, and C. eugenioides.
The clustered tomato diterpene synthase genes TPS18 and TPS21, and the monoterpene synthases TPS19 and TPS20, all classified as e/f [70] forms an orthogroup with N. cadamba, O. pumila, G. jasminoides, C. humblotiana, C. canephora, C. arabica, and C. eugenioides TPSs present in this cluster. The class c diterpene synthase gene TPS41 also shares orthogroups. Although the tomato cytochrome P450 have orthologs only in tomato, the MGCs from family FAM-1569 do contain cytochrome P450 genes in orthogroups that were exclusive to plants from the Rubiaceae family here analyzed. For FAM-1569, we identified both coexpression modules containing TPS and P450 genes from the same MGC in C. canephora, C. arabica and N. cadamba.
In tomato, the TPS18 gene synthesizes an unknown diterpene, TPS19 and TPS20 genes synthesize monoterpenes and the TPS21 gene synthesizes lycosantonolol. Those class e/f TPSs genes forms an orthogroup with N. cadamba, O. pumila, G. jasminoides, C. humblotiana, C. canephora, C. arabica, and C. eugenioides TPSs. The tomato class c diterpene synthase TPS41 gene also shares orthogroups with N. cadamba, O. pumila, C. humblotiana, C. canephora, C. arabica, and C. eugenioides TPSs. The tomato cytochrome P450 gene CYP71BN1 does not share orthogroups with plants of our study. Nevertheless, we observed that the cytochrome P450 genes from FAM-1569 MGCs share orthogroups with all plants in our study. Additionally, the CYP450 genes were identified as members of CYP71 and CYP76 clans, known to be involved in specialized diterpene metabolism [3]. The tomato cis-prenyl transferase gene (CPT1) has orthologs in N. cadamba and O. pumila, but such orthologs are distributed in other regions of their respective genomes. Future functional studies would help in the final understanding of the function of these genes.
The Rubiaceae genomes sampled in this study comprises three subfamilies strongly supported by previous phylogenies: Rubioideae (L. oblonga and O. pumila), Ixoroideae (Coffea spp. and G. jasminoides) and Cinchonoideae (N. cadamba) [6]. Taking account that members of the same subfamily should have more shared GCFs, plants of the Ixoroideae subfamily (Coffea spp. and G. jasminoides) had the major number of shared GCFs, which represent the most conserved metabolic gene clusters. Our results also suggest a major conservation among MGCs from Ixoroideae and Cinchonoideae subfamilies (here represented by N. cadamba), than Ixoroideae and Rubioideae subfamilies (here represented by L. oblonga and O. pumila). A total of 80 GCFs had representatives in all three subfamilies. When comparing with S. lycopersicum, we observed a higher number of conserved GCFs between S. lycopersicum and the Rubioideae subfamily, followed by S. lycopersicum and the Ixoroideae subfamily and finally, S. lycopersicum and the Cinchonoideae subfamily.
3.4. Cross-Species analysis of metabolic gene clusters unveils coexpression modules that contribute to set high confidence MGCs
A total of 1453 genes were found using both MGC discovery approaches. They were distributed in 217 clusters identified with PlantiSMASH and 211 clusters identified with PCF (supplementary Table S4). Coexpression analysis has been used to identify candidate genes associated with metabolic pathways. Genes that participate in the same metabolic pathway often display coordinated expression patterns when the environment changes [51], [68]. Thus, we conducted coexpression analysis using publicly available RNA-Seq experiments for Coffea arabica, Coffea canephora, Coffea eugenioides, Gardenia jasminoides, Ophiorrhiza pumila, and Neolamarckia cadamba.
Our analysis resulted in 19 coexpression modules across the five species (Figure s1-s6, Supplementary File 1). We examined whether genes within MGCs shared coexpression modules. Consequently, we considered MGCs with core genes in the same coexpression module as high-confidence MGCs.
In total, we identified 207 MGCs where at least three core metabolic genes were located in the same coexpression module. Of this total, 204 MGCs were also part of a gene cluster family, indicating conservation among other species in the study.
The coexpression analysis for the C. arabica dataset yielded two coexpression modules, with a total of 11 MGCs showing coexpression and conservation in other species. These MGCs were considered high-confidence (supplementary Table S6). For the C. canephora dataset, the coexpression analysis resulted in four coexpression modules, with 74 MGCs considered high-confidence (supplementary Table S7). The C. eugenioides dataset revealed four coexpression modules, with 9 MGCs considered high-confidence (supplementary Table S8). In the G. jasminoides dataset, the coexpression analysis identified three coexpression modules, and 11 MGCs were deemed high-confidence (supplementary Table S9). The O. pumila dataset yielded three modules in the coexpression analysis, with 17 MGCs considered high-confidence (supplementary Table S10). Lastly, the coexpression analysis for the N. cadamba dataset resulted in three coexpression modules, and a total of 82 MGCs were considered high-confidence (supplementary Table S11).
In addition to this, our cross-species analysis demonstrated the power of using coexpression modules for MGC identification. Genes within a metabolic pathway are often coexpressed, and finding these coexpression modules can provide strong evidence for the functional relevance of the predicted MGCs. In our study, we identified 207 high-confidence MGCs where at least three core metabolic genes were located in the same coexpression module. This approach not only enhances the confidence in MGC predictions but also provides a functional context to understand how these genes may work together in metabolic processes.
In conclusion, our analysis has successfully elucidated the complex landscape of MGCs across multiple plant species, paving the way for more targeted and in-depth studies in the future. The identification of coexpression modules also highlights the relevance of such cross-species comparative methods in unravelling potential functional associations and underlying genetic influences in metabolic pathways. One limitation of our study was the small amount of public transcriptome datasets suitable for co-expression analyses in the Rubiaceae family. While the number of libraries may be modest, this study serves as the initial systematic effort in co-expression analysis within this family laying groundwork for future. Our findings underscore the potential in harnessing this knowledge to enhance plant breeding programs and develop strategies for improved plant metabolic engineering.
Funding
This study was financed by CAPES-PrInt Program (process 88887.570702/2020–00). SMCL was financed in part by the Brazilian Federal Agency for Support and Evaluation of Graduate Education (CAPES)—Finance Code 001. NAPI Bioinformática from Fundação Araucária (# 66.2021). DSD research in coffee genomics and transcriptomics is supported by São Paulo State Research Foundation (FAPESP, grants #2016/10896–0 and #2018/08042–8) and by the Program for Support of New Faculty 2022/2023, at University of São Paulo. These funding agencies had no role in study design, the collection, analysis, and interpretation of data, or manuscript writing.
CRediT authorship contribution statement
Samara M. Correia de Lemos: Conceptualization, Methodology, Formal analysis, Data curation, Investigation, Writing – original draft, Writing – review & editing. Alexandre R. Paschoal: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing, Supervision. Romain Guyot: Methodology, Formal analysis, Investigation, Writing – review & editing, Supervision. Marnix Medema: Methodology, Formal analysis, Investigation, Writing – review & editing, Supervision. Douglas S. Domingues: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by resources supplied by the Center for Scientific Computing (NCC/GridUNESP) of the São Paulo State University (UNESP). The authors acknowledge the Wageningen University Bioinformatics Group at Wageningen University for providing computing resources that have contributed to the research results reported within this paper. The authors acknowledge the IRD itrop HPC (South Green Platform) at IRD Montpellier for providing HPC resources that have contributed to the research results reported within this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.5281/zenodo.8221450.
Appendix B. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Abu-Jamous B., Kelly S. Clust: automatic extraction of optimal co-expressed gene clusters from gene expression data. Genome Biol. 2018;19(1) doi: 10.1186/s13059-018-1536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adewole K.E., Attah A.F., Adebayo J.O. Morinda lucida Benth (Rubiaceae): a review of its ethnomedicine, phytochemistry and pharmacology. J Ethnopharmacol. 2021;276 doi: 10.1016/j.jep.2021.114055. [DOI] [PubMed] [Google Scholar]
- 3.Bathe U., Tissier A. Cytochrome P450 enzymes: a driving force of plant diterpene diversity. Phytochemistry. 2019;161:149–162. doi: 10.1016/j.phytochem.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bharadwaj R., Kumar S.R., Sharma A., Sathishkumar R. Plant metabolic gene clusters: evolution, organization, and their applications in synthetic biology. Front Plant Sci. 2021;12 doi: 10.3389/fpls.2021.697318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutanaev A.M., Moses T., Zi J., Nelson D.R., Mugford S.T., Peters R.J., Osbourn A. Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA. 2015;112(1):E81–E88. doi: 10.1073/pnas.1419547112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer B., Eriksson T. Time tree of Rubiaceae: phylogeny and dating the family, subfamilies and tribes. Int J Plant Sci. 2009;170:766–793. doi: 10.1086/599077. [DOI] [Google Scholar]
- 7.Brose J., Lau K.H., Dang T.T.T., Hamilton J.P., Martins L.D.V., Hamberger B., Hamberger B., Jiang J., O'Connor S.E., Buell C.R., 2021. The Mitragyna speciosa (Kratom) Genome: a resource for data-mining potent pharmaceuticals that impact human health. G3 (Bethesda). 11(4): jkab058. https://doi.org/10.1093/g3journal/jkab058. [DOI] [PMC free article] [PubMed]
- 8.Burge D. Conservation genomics and pollination biology of an endangered, edaphic-endemic, octoploid herb: El Dorado bedstraw (Galium californicum subsp. sierrae; Rubiaceae) PeerJ. 2020;8 doi: 10.7717/peerj.10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canales N.A., Pérez-Escobar O.A., Powell R.F., Töpel M., Kidner C., Nesbitt M., Maldonado C., Barnes C.J., Rønsted N., Przelomska N.A.S., Leitch I.J., Antonelli A. A highly contiguous, scaffold-level nuclear genome assembly for the fever tree (Cinchona pubescens Vahl) as a novel resource for Rubiaceae research. GigaByte. 2022;2022 doi: 10.46471/gigabyte.71. gigabyte71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chae L., Kim T., Nilo-Poyanco R., Rhee S.Y. Genomic signatures of specialized metabolism in plants. Science. 2014;344(6183):510–513. doi: 10.1126/science.1252076. [DOI] [PubMed] [Google Scholar]
- 11.Chen X., Liu F., Liu L., Qiu J., Fang D., Wang W., Zhang X., Ye C., Timko M.P., Zhu Q.H., Fan L., Xiao B. Characterization and evolution of gene clusters for terpenoid phytoalexin biosynthesis in tobacco. Planta. 2019;250(5):1687–1702. doi: 10.1007/s00425-019-03255-7. [DOI] [PubMed] [Google Scholar]
- 12.Dai B., Chen C., Liu Y., Liu L., Qaseem M.F., Wang J., Li H., Wu A.M. Physiological, biochemical, and transcriptomic responses of neolamarckia cadamba to aluminum stress. Int J Mol Sci. 2020;21(24):9624. doi: 10.3390/ijms21249624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denoeud F., Carretero-Paulet L., Dereeper A., Droc G., Guyot R., Pietrella M., Zheng C., Alberti A., Anthony F., Aprea G., Aury J.M., Bento P., Bernard M., Bocs S., Campa C., Cenci A., Combes M.C., Crouzillat D., Da Silva C., Daddiego L., De Bellis F., Dussert S., Garsmeur O., Gayraud T., Guignon V., Jahn K., Jamilloux V., Joët T., Labadie K., Lan T., Leclercq J., Lepelley M., Leroy T., Li L.T., Librado P., Lopez L., Muñoz A., Noel B., Pallavicini A., Perrotta G., Poncet V., Pot D., Priyono Rigoreau, Rouard M., Rozas M., Tranchant-Dubreuil J., VanBuren C., Zhang R., Andrade Q., Argout A.C., Bertrand X., de Kochko B., Graziosi A., Henry G., Jayarama R.J., Ming R., Nagai C., Rounsley S., Sankoff D., Giuliano G., Albert V.A., Wincker P., Lashermes P. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345(6201):1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 14.Domingues D.S., Oliveira L.S., Lemos S.M.C., Barros G.C.C., Ivamoto-Suzuki S.T. A bioinformatics tool for efficient retrieval of high-confidence terpene synthases (TPS) and application to the identification of TPS in Coffea and Quillaja. Methods Mol Biol. 2022;2469:43–53. doi: 10.1007/978-1-0716-2185-1_4. [DOI] [PubMed] [Google Scholar]
- 15.El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A., Sonnhammer E.L.L., Hirsh L., Paladin L., Piovesan D., Tosatto S.C.E., Finn R.D. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan P., Wang P., Lou Y., Leong B.J., Moore B.M., Schenck C.A., Combs R., Cao P., Brandizzi F., Shiu S., Last R.L. Evolution of a plant gene cluster in Solanaceae and emergence of metabolic diversity. eLife. 2020;9 doi: 10.7554/eLife.56717. e56717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist C.L.M., Booth T.J., van Wersch B., van Grieken L., Medema M.H., Chooi Y.H. cblaster: a remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform Adv. 2021;1(1) doi: 10.1093/bioadv/vbab016. vbab016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X.M., Wang Z.F., Zhang Y., Wang R.J. Chromosomal-level assembly of the Leptodermis oblonga (Rubiaceae) genome and its phylogenetic implications. Genomics. 2021;113(5):3072–3082. doi: 10.1016/j.ygeno.2021.07.012. [DOI] [PubMed] [Google Scholar]
- 20.Hawkins C., Ginzburg D., Zhao K., Dwyer W., Xue B., Xu A., Rice S., Cole B., Paley S., Karp P., Rhee S.Y. Plant Metabolic Network 15: A resource of genome-wide metabolism databases for 126 plants and algae. J Integr Plant Biol. 2021;63(11):1888–1905. doi: 10.1111/jipb.13163. [DOI] [PubMed] [Google Scholar]
- 21.Hosmani P.S., Flores-Gonzalez M., van de Geest H., Maumus F., Bakker L.V., Schijlen E., van Haarst J., Cordewener J., Sanchez-Perez G., Peters S., Fei Z., Giovannoni J.J., Mueller L.A., Saha S. An improved de novo assembly and annotation of the tomato reference genome using single-molecule sequencing, Hi-C proximity ligation and optical maps. bioRxiv. 2019 doi: 10.1101/767764. [DOI] [Google Scholar]
- 22.Julca I., Mutwil-Anderwald D., Manoj V., Khan Z., Lai S.K., Yang L.K., Beh I.T., Dziekan J., Lim Y.P., Lim S.K., Low Y.W., Lam Y.I., Tjia S., Mu Y., Tan Q.W., Nuc P., Choo L.M., Khew G., Shining L., Kam A., Tam J.P., Bozdech Z., Schmidt M., Usadel B., Kanagasundaram Y., Alseekh S., Fernie A., Li H.Y., Mutwil M. Genomic, transcriptomic, and metabolomic analysis of Oldenlandia corymbosa reveals the biosynthesis and mode of action of anti-cancer metabolites. J Integr Plant Biol. 2023;65(6):1442–1466. doi: 10.1111/jipb.13469. [DOI] [PubMed] [Google Scholar]
- 23.Karp P.D., Paley S., Krummenacker M., Kothari A., Wannemuehler M.J., Phillips G.J. Pathway Tools Management of Pathway/Genome Data for Microbial Communities. Front Bioinform. 2022;2 doi: 10.3389/fbinf.2022.869150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kautsar S.A., Suarez Duran H.G., Blin K., Osbourn A., Medema M.H. PlantiSMASH: automated identification, annotation and expression analysis of plant biosynthetic gene clusters. Nucleic Acids Res. 2017;45(W1):W55–W63. doi: 10.1093/nar/gkx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kautsar S.A., Blin K., Shaw S., Weber T., Medema M.H. BiG-FAM: the biosynthetic gene cluster families database. Nucleic Acids Res. 2021;49(D1):D490–D497. doi: 10.1093/nar/gkaa812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kessler A., Kalske A. Plant Secondary Metabolite Diversity and Species Interactions. Annu Rev Ecol, Evol, Syst. 2018;49:115–138. doi: 10.1146/annurev-ecolsys-110617-062406. [DOI] [Google Scholar]
- 27.Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau K.H., Bhat W.W., Hamilton J.P., Wood J.C., Vaillancourt B., Wiegert-Rininger K., Newton L., Hamberger B., Holmes D., Hamberger B., Buell C.R. Genome assembly of Chiococca alba uncovers key enzymes involved in the biosynthesis of unusual terpenoids. DNA Res. 2020;27(3) doi: 10.1093/dnares/dsaa013. dsaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li C.Y., Yang L., Liu Y., Xu Z.G., Gao J., Huang Y.B., Xu J.J., Fan H., Kong Y., Wei Y.K., Hu W.L., Wang L.J., Zhao Q., Hu Y.H., Zhang Y.J., Martin C., Chen X.Y. The sage genome provides insight into the evolutionary dynamics of diterpene biosynthesis gene cluster in plants. Cell Rep. 2022;40(7) doi: 10.1016/j.celrep.2022.111236. [DOI] [PubMed] [Google Scholar]
- 30.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 31.Liu C., Smit S.J., Dang J., Zhou P., Godden G.T., Jiang Z., Liu W., Liu L., Lin W., Duan J., Wu Q., Lichman B.R. A chromosome-level genome assembly reveals that a bipartite gene cluster formed via an inverted duplication controls monoterpenoid biosynthesis in Schizonepeta tenuifolia. Mol Plant. 2023;16(3):533–548. doi: 10.1016/j.molp.2023.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Maeda H.A., Fernie A.R. Evolutionary History of Plant Metabolism. Annu Rev Plant Biol. 2021;72:185–216. doi: 10.1146/annurev-arplant-080620-031054. [DOI] [PubMed] [Google Scholar]
- 33.Martins D., Nunez C.V. Secondary metabolites from Rubiaceae species. Molecules. 2015;20(7):13422–13495. doi: 10.3390/molecules200713422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuba Y., Nguyen T.T., Wiegert K., Falara V., Gonzales-Vigil E., Leong B., Schäfer P., Kudrna D., Wing R.A., Bolger A.M., Usadel B., Tissier A., Fernie A.R., Barry C.S., Pichersky E. Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell. 2013;25(6):2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuba Y., Zi J., Jones A.D., Peters R.J., Pichersky E. Biosynthesis of the diterpenoid lycosantalonol via nerylneryl diphosphate in Solanum lycopersicum. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medema M.H., Osbourn A. Computational genomic identification and functional reconstitution of plant natural product biosynthetic pathways. Nat Prod Rep. 2016;33(8):951–962. doi: 10.1039/c6np00035e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohite O.S., Lloyd C.J., Monk J.M., Weber T., Palsson B.O. Pangenome analysis of Enterobacteria reveals richness of secondary metabolite gene clusters and their associated gene sets. Synth Syst Biotechnol. 2022;7(3):900–910. doi: 10.1016/j.synbio.2022.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarro-Muñoz J.C., Selem-Mojica N., Mullowney M.W., Kautsar S.A., Tryon J.H., Parkinson E.I., De Los Santos E.L.C., Yeong M., Cruz-Morales P., Abubucker S., Roeters A., Lokhorst W., Fernandez-Guerra A., Cappelini L.T.D., Goering A.W., Thomson R.J., Metcalf W.W., Kelleher N.L., Barona-Gomez F., Medema M.H. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2020;16(1):60–68. doi: 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan Y., Zhao X., Wang Y., Tan J., Chen D.X. Metabolomics integrated with transcriptomics reveals the distribution of iridoid and crocin metabolic flux in Gardenia jasminoides Ellis. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0256802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perrois C., Strickler S.R., Mathieu G., Lepelley M., Bedon L., Michaux S., Husson J., Mueller L., Privat I. Differential regulation of caffeine metabolism in Coffea arabica (Arabica) and Coffea canephora (Robusta) Planta. 2015;241(1):179–191. doi: 10.1007/s00425-014-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pichersky E., Lewinsohn E. Convergent evolution in plant specialized metabolism. Annu Rev Plant Biol. 2011;62:549–566. doi: 10.1146/annurev-arplant-042110-103814. [DOI] [PubMed] [Google Scholar]
- 42.Polturak G., Liu Z., Osbourn A. New and emerging concepts in the evolution and function of plant biosynthetic gene clusters. Curr Opin Green Sustain Chem. 2022;33 doi: 10.1016/j.cogsc.2021.100568. [DOI] [Google Scholar]
- 43.Pucker B., Reiher F., Schilbert H.M. Automatic Identification of Players in the Flavonoid Biosynthesis with Application on the Biomedicinal Plant Croton tiglium. Plants. 2020;9(9):1103. doi: 10.3390/plants9091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabara R.C., Kudithipudi C., Timko M.P. Identification of Terpene-Related Biosynthetic Gene Clusters in Tobacco through Computational-Based Genomic, Transcriptomic, and Metabolic Analyses. Agronomy. 2023;13(6):1632. doi: 10.3390/agronomy13061632. [DOI] [Google Scholar]
- 45.Raharimalala N., Rombauts S., McCarthy A., Garavito A., Orozco-Arias S., Bellanger L., Morales-Correa A.Y., Froger S., Michaux S., Berry V., Metairon S., Fournier C., Lepelley M., Mueller L., Couturon E., Hamon P., Rakotomalala J.J., Descombes P., Guyot R., Crouzillat D. The absence of the caffeine synthase gene is involved in the naturally decaffeinated status of Coffea humblotiana, a wild species from Comoro archipelago. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-87419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rai A., Hirakawa H., Nakabayashi R., Kikuchi S., Hayashi K., Rai M., Tsugawa H., Nakaya T., Mori T., Nagasaki H., Fukushi R., Kusuya Y., Takahashi H., Uchiyama H., Toyoda A., Hikosaka S., Goto E., Saito K., Yamazaki M. Chromosome-level genome assembly of Ophiorrhiza pumila reveals the evolution of camptothecin biosynthesis. Nat Commun. 2021;12(1) doi: 10.1038/s41467-020-20508-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rieseberg T.P., Dadras A., Fürst-Jansen J.M.R., et al. Crossroads in the evolution of plant specialized metabolism. Semin Cell Dev Biol. 2023;134:37–58. doi: 10.1016/j.semcdb.2022.03.004. ISSN 1084-9521. [DOI] [PubMed] [Google Scholar]
- 48.Robey M.T., Caesar L.K., Drott M.T., Keller N.P., Kelleher N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc Natl Acad Sci USA. 2021;118(19) doi: 10.1073/pnas.2020230118. e2020230118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schläpfer P., Zhang P., Wang C., Kim T., Banf M., Chae L., Dreher K., Chavali A.K., Nilo-Poyanco R., Bernard T., Kahn D., Rhee S.Y. Genome-Wide Prediction of Metabolic Enzymes, Pathways, and Gene Clusters in Plants. Plant Physiol. 2017;173(4):2041–2059. doi: 10.1104/pp.16.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 2016;9 doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh K.S., van der Hooft J.J.J., van Wees S.C.M., Medema M.H. Integrative omics approaches for biosynthetic pathway discovery in plants. Nat Prod Rep. 2022;39(9):1876–1896. doi: 10.1039/d2np00032f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi M., Gong H., Cui L., Wang Q., Wang C., Wang Y., Kai G. Targeted metabolic engineering of committed steps improves anti-cancer drug camptothecin production in Ophiorrhiza pumila hairy roots. Ind Crops Prod. 2020;148 doi: 10.1016/j.indcrop.2020.112277. ISSN 0926-6690. [DOI] [Google Scholar]
- 53.Smit S.J., Lichman B.R. Plant biosynthetic gene clusters in the context of metabolic evolution. Nat Prod Rep. 2022;39(7):1465–1482. doi: 10.1039/d2np00005a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Srivastav V.K., Egbuna C., Tiwari M. Phytochemicals as Lead Compounds for New Drug Discovery. Elsevier; 2020. Chapter 1 - Plant secondary metabolites as lead compounds for the production of potent drugs; pp. 3–14. ISBN 9780128178904. [DOI] [Google Scholar]
- 55.Stavrinides A.K., Dussert S., Combes M.C., Fock-Bastide I., Severac D., Minier J., Bastos-Siqueira A., Demolombe V., Hem S., Lashermes P., Joët T. Seed comparative genomics in three coffee species identify desiccation tolerance mechanisms in intermediate seeds. J Exp Bot. 2020;71(4):1418–1433. doi: 10.1093/jxb/erz508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J., Lu F., Luo Y., Bie L., Xu L., Wang Y. OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 2023;51(W1):W397–W403. doi: 10.1093/nar/gkad313. PMID: 37114999; PMCID: PMC10320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terlouw B.R., Blin K., Navarro-Muñoz J.C., Avalon N.E., Chevrette M.G., Egbert S., Lee S., Meijer D., Recchia M.J.J., Reitz Z.L., van Santen J.A., Selem-Mojica N., Tørring T., Zaroubi L., Alanjary M., Aleti G., Aguilar C., Al-Salihi S.A.A., Augustijn H.E., Avelar-Rivas J.A., Avitia-Domínguez L.A., Barona-Gómez F., Bernaldo-Agüero J., Bielinski V.A., Biermann F., Booth T.J., Carrion Bravo V.J., Castelo-Branco R., Chagas F.O., Cruz-Morales P., Du C., Duncan K.R., Gavriilidou A., Gayrard D., Gutiérrez-García K., Haslinger K., Helfrich E.J.N., van der Hooft J.J.J., Jati A.P., Kalkreuter E., Kalyvas N., Kang K.B., Kautsar S., Kim W., Kunjapur A.M., Li Y.X., Lin G.M., Loureiro C., Louwen J.J.R., Louwen N.L.L., Lund G., Parra J., Philmus B., Pourmohsenin B., Pronk L.J.U., Rego A., Rex D.A.B., Robinson S., Rosas-Becerra L.R., Roxborough E.T., Schorn M.A., Scobie D.J., Singh K.S., Sokolova N., Tang X., Udwary D., Vigneshwari A., Vind K., Vromans S.P.J.M., Waschulin V., Williams S.E., Winter J.M., Witte T.E., Xie H., Yang D., Yu J., Zdouc M., Zhong Z., Collemare J., Linington R.G., Weber T., Medema M.H. MIBiG 3.0: a community-driven effort to annotate experimentally validated biosynthetic gene clusters. Nucleic Acids Res. 2023;51(D1):D603–D610. doi: 10.1093/nar/gkac1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tohge T., Fernie A.R. Co-regulation of clustered and neo-functionalized genes in plant-specialized metabolism. Plants (Basel) 2020;9(5):622. doi: 10.3390/plants9050622. PMID: 32414181; PMCID: PMC7285293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Twaij B.M., Hasan M.N. Bioactive secondary metabolites from plant sources: types, synthesis, and their therapeutic uses. Int J Plant Biol. 2022;13(1):4–14. doi: 10.3390/ijpb13010003. 2022; [DOI] [Google Scholar]
- 60.Udomsom N., Rai A., Suzuki H., Okuyama J., Imai R., Mori T., Nakabayashi R., Saito K., Yamazaki M. Function of AP2/ERF Transcription Factors Involved in the Regulation of Specialized Metabolism in Ophiorrhiza pumila Revealed by Transcriptomics and Metabolomics. Front Plant Sci. 2016;7:1861. doi: 10.3389/fpls.2016.01861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Xu S., Mei Y., Cai S., Gu Y., Sun M., Liang Z., Xiao Y., Zhang M., Yang S. A high-quality genome assembly of Morinda officinalis, a famous native southern herb in the Lingnan region of southern China. Hortic Res. 2021;8 doi: 10.1038/s41438-021-00551-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang P., Schumacher A.M., Shiu S.H. Computational prediction of plant metabolic pathways. Curr Opin Plant Biol. 2022;66 doi: 10.1016/j.pbi.2021.102171. [DOI] [PubMed] [Google Scholar]
- 63.Wingett S.W., Andrews S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research. 2018;7:1338. doi: 10.12688/f1000research.15931.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu S., Malaco Morotti A.L., Wang S., Wang Y., Xu X., Chen J., Wang G., Tatsis E.C. Convergent gene clusters underpin hyperforin biosynthesis in St John's wort. N Phytol. 2022;235(2):646–661. doi: 10.1111/nph.18138. [DOI] [PubMed] [Google Scholar]
- 65.Xu Z., Pu X., Gao R., Demurtas O.C., Fleck S.J., Richter M., He C., Ji A., Sun W., Kong J., Hu K., Ren F., Song J., Wang Z., Gao T., Xiong C., Yu H., Xin T., Albert V.A., Giuliano G., Chen S., Song J. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020;18(1) doi: 10.1186/s12915-020-00795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang X., Zhang L., Guo X., Xu J., Zhang K., Yang Y., Yang Y., Jian Y., Dong D., Huang S., Cheng F., Li G. The gap-free potato genome assembly reveals large tandem gene clusters of agronomical importance in highly repeated genomic regions. Mol Plant. 2023;16(2):314–317. doi: 10.1016/j.molp.2022.12.010. [DOI] [PubMed] [Google Scholar]
- 67.Zhan C., Shen S., Yang C., Liu Z., Fernie A.R., Graham I.A., Luo J. Plant metabolic gene clusters in the multi-omics era. Trends Plant Sci. 2022;27(10):981–1001. doi: 10.1016/j.tplants.2022.03.002. [DOI] [PubMed] [Google Scholar]
- 68.Zhao K., Rhee S.Y. Omics-guided metabolic pathway discovery in plants: Resources, approaches, and opportunities. Curr Opin Plant Biol. 2022;67 doi: 10.1016/j.pbi.2022.102222. [DOI] [PubMed] [Google Scholar]
- 69.Zhao X., Hu X., OuYang K., Yang J., Que Q., Long J., Zhang J., Zhang T., Wang X., Gao J., Hu X., Yang S., Zhang L., Li S., Gao W., Li B., Jiang W., Nielsen E., Chen X., Peng C. Chromosome-level assembly of the Neolamarckia cadamba genome provides insights into the evolution of cadambine biosynthesis. Plant J. 2022;109(4):891–908. doi: 10.1111/tpj.15600. [DOI] [PubMed] [Google Scholar]
- 70.Zhou F., Pichersky E. The complete functional characterisation of the terpene synthase family in tomato. N Phytol. 2020;226(5):1341–1360. doi: 10.1111/nph.16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material