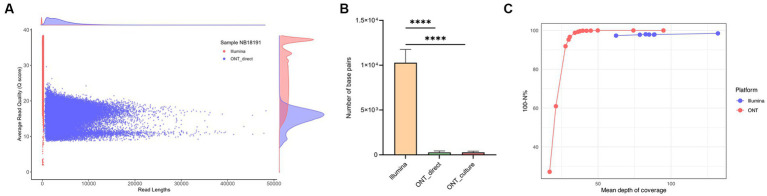
Figure 4.
Coverage and genome quality across the reference, comparison of culture-derived Illumina reads with culture-derived and culture-independent reads generated with Nanopore adaptive sampling (NAS). (A) Distribution of read lengths and average quality (Q values) for Illumina and culture-independent data generated with NAS for sample NB18191. (B) The number of bases with no coverage (depth = 0) was compared between the three groups. The Illumina dataset showed the highest number of bases with no coverage with 0.24% (M = 10,279 positions, SD = 1,369.24), followed by the two ONT datasets with similar results and 0.006% of the reference genome not being covered (ONT culture M = 275 positions, SD = 126.2 and ONT direct M = 271 positions, SD = 148.73) (****p < 0.0001). (C) Comparison of mean depth of coverage for M. bovis genomes and their quality at single position level. The number of positions with a sequencing depth below 20X (N%) was compared with corresponding mean depth of coverage over the entire genome.