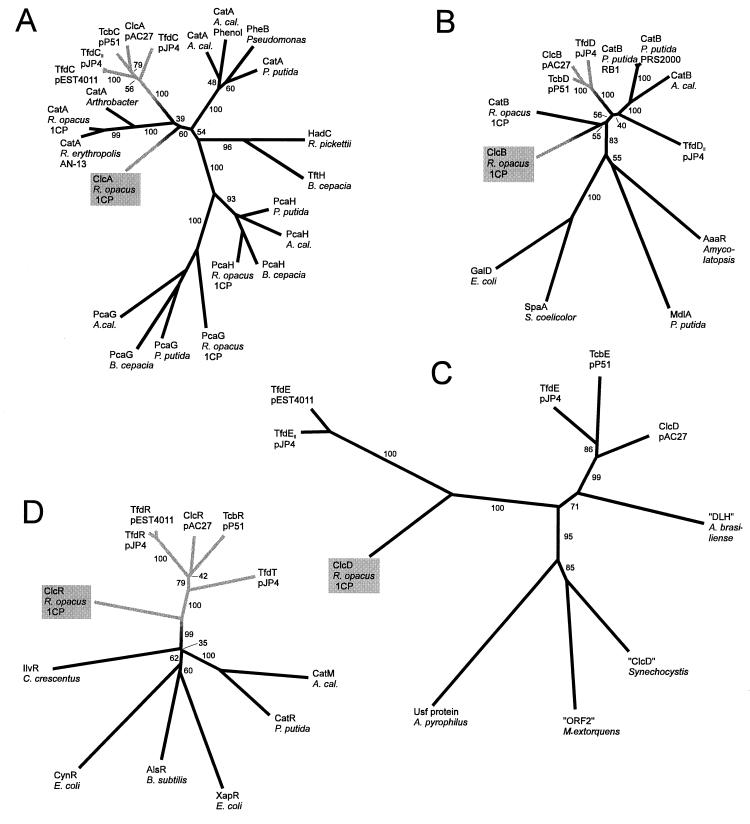
FIG. 8.
Dendrograms illustrating sequence similarities between proteins involved in catechol and chlorocatechol catabolism. (A) Catechol and chlorocatechol 1,2-dioxygenases; (B) muconate and chloromuconate cycloisomerases; (C) dienelactone hydrolases; (D) LysR-type regulatory proteins. The dendrograms were calculated by the PHYLIP program package (19) based on CLUSTAL alignments of the sequences in Fig. 4 to 7 and the respective outgroup sequences. For the sequences used in Fig. 4 to 7, the resulting alignments were very similar but, close to some gaps, not quite identical to those shown. Numbers on the branches indicate the frequency (percentage) with which the corresponding cluster occurred during bootstrap analyses. They show that the affiliation of a certain protein to one of the major groups could be deduced with great confidence. The exact branching pattern of these major groups, in contrast, as well as the branching within the groups is not certain, as indicated by low bootstrap numbers. The dendrograms for cycloisomerases and dienelactone hydrolases were calculated from the complete alignments (with the exception that the amino acids in positions 3 to 38 of “ORF2” of Methylobacterium extorquens were omitted). For the dioxygenases, only positions 140 to 258 of the alignment in Fig. 6, and for the regulators, only positions 1 to 217 of the alignment in Fig. 5, were considered. All dendrograms are drawn to the same scale with respect to evolutionary distance (in centimeters per unit branch length). Gray branches represent enzymes which are specific for chlorocatechol catabolism; the position of the transition from black to gray was chosen arbitrarily. Functions and accessions numbers of the proteins used as outgroups: PcaG and PcaH, α and β subunits of protocatechuate 3,4-dioxygenases, of A. calcoaceticus (A. cal.) (M33798), Burkholderia cepacia (M30791), P. putida (L14836) and R. opacus (AF003947) TftH, hydroxyquinol 1,2-dioxygenase, of B. cepacia (U19883); HadC, hydroxyquinol 1,2-dioxygenase, of Ralstonia pickettii (D86544); GalD, galactonate dehydratase, of E. coli (L10328 and U19577); SpaA, protein possibly involved in the metabolism of signalling lactones, of Streptomyces coelicolor (X94190); MdlA, mandelate racemase, of P. putida (J05293); AaaR, N-acyl amino acid racemase, of Amycolatopsis sp. (D30738); IlvR, regulator of isoleucine and valine synthesis, of Caulobacter crescentus (L24392); CynR, regulator for cyanate detoxification, of E. coli (M93053); AlsR, acetoin synthesis regulator, of Bacillus subtilis (L04470); XapR, xanthosine catabolic regulator, of E. coli (X73828); Usf protein, hypothetical protein, of Aquifex pyrophilus (U17575); “ORF2,” hypothetical protein, of M. extorquens (U72662); “ClcD,” hypothetical protein homologous to dienelactone hydrolases, of Synechocystis sp. PCC6803 (D90904); “DLH,” hypothetical protein homologous to dienelactone hydrolases, of Azospirillum brasiliense (X67216).